In vivo mapping of the mouse Galnt3-specific O-glycoproteome
- PMID: 39098533
- PMCID: PMC11402288
- DOI: 10.1016/j.jbc.2024.107628
In vivo mapping of the mouse Galnt3-specific O-glycoproteome
Abstract
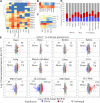
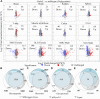
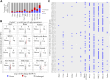
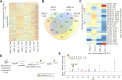
The UDP-N-acetylgalactosamine polypeptide:N-acetylgalactosaminyltransferase (GalNAc-T) family of enzymes initiates O-linked glycosylation by catalyzing the addition of the first GalNAc sugar to serine or threonine on proteins destined to be membrane-bound or secreted. Defects in individual isoforms of the GalNAc-T family can lead to certain congenital disorders of glycosylation (CDG). The polypeptide N-acetylgalactosaminyltransferase 3 (GALNT)3-CDG, is caused by mutations in GALNT3, resulting in hyperphosphatemic familial tumoral calcinosis due to impaired glycosylation of the phosphate-regulating hormone fibroblast growth factor 23 (FGF23) within osteocytes of the bone. Patients with hyperphosphatemia present altered bone density, abnormal tooth structure, and calcified masses throughout the body. It is therefore important to identify all potential substrates of GalNAc-T3 throughout the body to understand the complex disease phenotypes. Here, we compared the Galnt3-/- mouse model, which partially phenocopies GALNT3-CDG, with WT mice and used a multicomponent approach using chemoenzymatic conditions, a product-dependent method constructed using EThcD triggered scans in a mass spectrometry workflow, quantitative O-glycoproteomics, and global proteomics to identify 663 Galnt3-specific O-glycosites from 269 glycoproteins across multiple tissues. Consistent with the mouse and human phenotypes, functional networks of glycoproteins that contain GalNAc-T3-specific O-glycosites involved in skeletal morphology, mineral level maintenance, and hemostasis were identified. This library of in vivo GalNAc-T3-specific substrate proteins and O-glycosites will serve as a valuable resource to understand the functional implications of O-glycosylation and to unravel the underlying causes of complex human GALNT3-CDG phenotypes.
Keywords: GalNAc-T; Galnt; O-GalNAc; O-glycosylation; glycopeptides; glycosyltransferase; hyperphosphatemic familial tumoral calcinosis (HFTC); mass spectrometry.
Published by Elsevier Inc.
Conflict of interest statement
Conflict of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures



Similar articles
-
Loss of the disease-associated glycosyltransferase Galnt3 alters Muc10 glycosylation and the composition of the oral microbiome.J Biol Chem. 2020 Jan 31;295(5):1411-1425. doi: 10.1074/jbc.RA119.009807. Epub 2019 Dec 27. J Biol Chem. 2020. PMID: 31882545 Free PMC article.
-
Quantitative mapping of the in vivo O-GalNAc glycoproteome in mouse tissues identifies GalNAc-T2 O-glycosites in metabolic disorder.Proc Natl Acad Sci U S A. 2023 Oct 24;120(43):e2303703120. doi: 10.1073/pnas.2303703120. Epub 2023 Oct 20. Proc Natl Acad Sci U S A. 2023. PMID: 37862385 Free PMC article.
-
Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression.Endocrinology. 2009 Jun;150(6):2543-50. doi: 10.1210/en.2008-0877. Epub 2009 Feb 12. Endocrinology. 2009. PMID: 19213845 Free PMC article.
-
Polypeptide N-acetylgalactosaminyltransferase-Associated Phenotypes in Mammals.Molecules. 2021 Sep 10;26(18):5504. doi: 10.3390/molecules26185504. Molecules. 2021. PMID: 34576978 Free PMC article. Review.
-
Miscellaneous non-inflammatory musculoskeletal conditions. Hyperphosphatemic familial tumoral calcinosis (FGF23, GALNT3 and αKlotho).Best Pract Res Clin Rheumatol. 2011 Oct;25(5):735-47. doi: 10.1016/j.berh.2011.10.020. Best Pract Res Clin Rheumatol. 2011. PMID: 22142751 Free PMC article. Review.
References
-
- Freeze H.H. Genetic defects in the human glycome. Nat. Rev. Genet. 2006;7:537–551. - PubMed
-
- Topaz O., Shurman D.L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., et al. Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 2004;36:579–581. - PubMed
-
- Ichikawa S., Sorenson A.H., Austin A.M., Mackenzie D.S., Fritz T.A., Moh A., et al. Ablation of the Galnt3 gene leads to low-circulating intact fibroblast growth factor 23 (Fgf23) concentrations and hyperphosphatemia despite increased Fgf23 expression. Endocrinology. 2009;150:2543–2550. - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

