The genome of the colonial hydroid Hydractinia reveals that their stem cells use a toolkit of evolutionarily shared genes with all animals
- PMID: 38508693
- PMCID: PMC11067881
- DOI: 10.1101/gr.278382.123
The genome of the colonial hydroid Hydractinia reveals that their stem cells use a toolkit of evolutionarily shared genes with all animals
Abstract
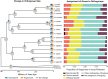
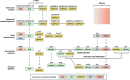
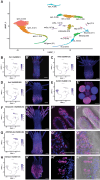
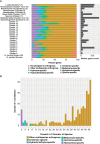
Hydractinia is a colonial marine hydroid that shows remarkable biological properties, including the capacity to regenerate its entire body throughout its lifetime, a process made possible by its adult migratory stem cells, known as i-cells. Here, we provide an in-depth characterization of the genomic structure and gene content of two Hydractinia species, Hydractinia symbiolongicarpus and Hydractinia echinata, placing them in a comparative evolutionary framework with other cnidarian genomes. We also generated and annotated a single-cell transcriptomic atlas for adult male H. symbiolongicarpus and identified cell-type markers for all major cell types, including key i-cell markers. Orthology analyses based on the markers revealed that Hydractinia's i-cells are highly enriched in genes that are widely shared amongst animals, a striking finding given that Hydractinia has a higher proportion of phylum-specific genes than any of the other 41 animals in our orthology analysis. These results indicate that Hydractinia's stem cells and early progenitor cells may use a toolkit shared with all animals, making it a promising model organism for future exploration of stem cell biology and regenerative medicine. The genomic and transcriptomic resources for Hydractinia presented here will enable further studies of their regenerative capacity, colonial morphology, and ability to distinguish self from nonself.
© 2024 Schnitzler et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Update of
-
The genome of the colonial hydroid Hydractinia reveals their stem cells utilize a toolkit of evolutionarily shared genes with all animals.bioRxiv [Preprint]. 2023 Aug 27:2023.08.25.554815. doi: 10.1101/2023.08.25.554815. bioRxiv. 2023. Update in: Genome Res. 2024 Apr 25;34(3):498-513. doi: 10.1101/gr.278382.123. PMID: 37786714 Free PMC article. Updated. Preprint.
Similar articles
-
The genome of the colonial hydroid Hydractinia reveals their stem cells utilize a toolkit of evolutionarily shared genes with all animals.bioRxiv [Preprint]. 2023 Aug 27:2023.08.25.554815. doi: 10.1101/2023.08.25.554815. bioRxiv. 2023. Update in: Genome Res. 2024 Apr 25;34(3):498-513. doi: 10.1101/gr.278382.123. PMID: 37786714 Free PMC article. Updated. Preprint.
-
The transcriptome of the colonial marine hydroid Hydractinia echinata.FEBS J. 2010 Jan;277(1):197-209. doi: 10.1111/j.1742-4658.2009.07474.x. Epub 2009 Dec 3. FEBS J. 2010. PMID: 19961538
-
GNL3 is an evolutionarily conserved stem cell gene influencing cell proliferation, animal growth and regeneration in the hydrozoan Hydractinia.Open Biol. 2022 Sep;12(9):220120. doi: 10.1098/rsob.220120. Epub 2022 Sep 7. Open Biol. 2022. PMID: 36069077 Free PMC article.
-
The Hydractinia allorecognition system.Immunogenetics. 2022 Feb;74(1):27-34. doi: 10.1007/s00251-021-01233-6. Epub 2021 Nov 13. Immunogenetics. 2022. PMID: 34773127 Review.
-
The hydroid Hydractinia: a versatile, informative cnidarian representative.Bioessays. 2001 Oct;23(10):963-71. doi: 10.1002/bies.1137. Bioessays. 2001. PMID: 11598963 Review.
Cited by
-
Regeneration in the absence of canonical neoblasts in an early branching flatworm.bioRxiv [Preprint]. 2024 May 28:2024.05.24.595708. doi: 10.1101/2024.05.24.595708. bioRxiv. 2024. PMID: 38853907 Free PMC article. Preprint.
References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
