This is a preprint.
IgM N-glycosylation correlates with COVID-19 severity and rate of complement deposition
- PMID: 37398192
- PMCID: PMC10312960
- DOI: 10.21203/rs.3.rs-2939468/v1
IgM N-glycosylation correlates with COVID-19 severity and rate of complement deposition
Update in
-
IgM N-glycosylation correlates with COVID-19 severity and rate of complement deposition.Nat Commun. 2024 Jan 9;15(1):404. doi: 10.1038/s41467-023-44211-0. Nat Commun. 2024. PMID: 38195739 Free PMC article.
Abstract
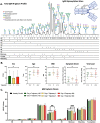
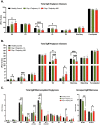
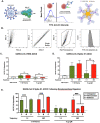
The glycosylation of IgG plays a critical role during human SARS-CoV-2, activating immune cells and inducing cytokine production. However, the role of IgM N-glycosylation has not been studied during acute viral infection in humans. In vitro evidence suggests that the glycosylation of IgM inhibits T cell proliferation and alters complement activation rates. The analysis of IgM N-glycosylation from healthy controls and hospitalized COVID-19 patients reveals that mannosylation and sialyation levels associate with COVID-19 severity. Specifically, we find increased di- and tri-sialylated glycans and altered mannose glycans in total serum IgM in severe COVID-19 patients when compared to moderate COVID-19 patients. This is in direct contrast with the decrease of sialic acid found on the serum IgG from the same cohorts. Moreover, the degree of mannosylation and sialylation correlated significantly with markers of disease severity: D-dimer, BUN, creatinine, potassium, and early anti-COVID-19 amounts of IgG, IgA, and IgM. Further, IL-16 and IL-18 cytokines showed similar trends with the amount of mannose and sialic acid present on IgM, implicating these cytokines' potential to impact glycosyltransferase expression during IgM production. When examining PBMC mRNA transcripts, we observe a decrease in the expression of Golgi mannosidases that correlates with the overall reduction in mannose processing we detect in the IgM N-glycosylation profile. Importantly, we found that IgM contains alpha-2,3 linked sialic acids in addition to the previously reported alpha-2,6 linkage. We also report that antigen-specific IgM antibody-dependent complement deposition is elevated in severe COVID-19 patients. Taken together, this work links the immunoglobulin M N-glycosylation with COVID-19 severity and highlights the need to understand the connection between IgM glycosylation and downstream immune function during human disease.
Keywords: COVID-19; Complement Deposition; Glycomics; IgM N-glycan; Immunoglobulin M; SARS-CoV-2.
Conflict of interest statement
Declarations Competing Interests The authors have no competing interests to declare.
Figures


Similar articles
-
IgM N-glycosylation correlates with COVID-19 severity and rate of complement deposition.Nat Commun. 2024 Jan 9;15(1):404. doi: 10.1038/s41467-023-44211-0. Nat Commun. 2024. PMID: 38195739 Free PMC article.
-
A new multiplex SARS-CoV-2 antigen microarray showed correlation of IgG, IgA, and IgM antibodies from patients with COVID-19 disease severity and maintenance of relative IgA and IgM antigen binding over time.PLoS One. 2023 Mar 30;18(3):e0283537. doi: 10.1371/journal.pone.0283537. eCollection 2023. PLoS One. 2023. PMID: 36996259 Free PMC article.
-
α2,6-Sialylation Is Upregulated in Severe COVID-19, Implicating the Complement Cascade.ACS Infect Dis. 2022 Nov 11;8(11):2348-2361. doi: 10.1021/acsinfecdis.2c00421. Epub 2022 Oct 11. ACS Infect Dis. 2022. PMID: 36219583 Free PMC article.
-
Sialylation as an Important Regulator of Antibody Function.Front Immunol. 2022 Apr 7;13:818736. doi: 10.3389/fimmu.2022.818736. eCollection 2022. Front Immunol. 2022. PMID: 35464485 Free PMC article. Review.
-
IgG N-glycans.Adv Clin Chem. 2021;105:1-47. doi: 10.1016/bs.acc.2021.02.001. Epub 2021 Mar 18. Adv Clin Chem. 2021. PMID: 34809825 Review.
References
Publication types
Grants and funding
- U19 AI090023/AI/NIAID NIH HHS/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- UL1 TR004419/TR/NCATS NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

