Antibody evasion properties of SARS-CoV-2 Omicron sublineages
- PMID: 35240676
- PMCID: PMC9021018
- DOI: 10.1038/s41586-022-04594-4
Antibody evasion properties of SARS-CoV-2 Omicron sublineages
Abstract
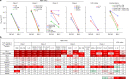
The identification of the Omicron (B.1.1.529.1 or BA.1) variant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in Botswana in November 20211 immediately caused concern owing to the number of alterations in the spike glycoprotein that could lead to antibody evasion. We2 and others3-6 recently reported results confirming such a concern. Continuing surveillance of the evolution of Omicron has since revealed the rise in prevalence of two sublineages, BA.1 with an R346K alteration (BA.1+R346K, also known as BA.1.1) and B.1.1.529.2 (BA.2), with the latter containing 8 unique spike alterations and lacking 13 spike alterations found in BA.1. Here we extended our studies to include antigenic characterization of these new sublineages. Polyclonal sera from patients infected by wild-type SARS-CoV-2 or recipients of current mRNA vaccines showed a substantial loss in neutralizing activity against both BA.1+R346K and BA.2, with drops comparable to that already reported for BA.1 (refs. 2,3,5,6). These findings indicate that these three sublineages of Omicron are antigenically equidistant from the wild-type SARS-CoV-2 and thus similarly threaten the efficacies of current vaccines. BA.2 also exhibited marked resistance to 17 of 19 neutralizing monoclonal antibodies tested, including S309 (sotrovimab)7, which had retained appreciable activity against BA.1 and BA.1+R346K (refs. 2-4,6). This finding shows that no authorized monoclonal antibody therapy could adequately cover all sublineages of the Omicron variant, except for the recently authorized LY-CoV1404 (bebtelovimab).
© 2022. The Author(s).
Conflict of interest statement
S.I., Lihong Liu, J.Y., Yaoxing Huang and D.D.H. are inventors on patent applications (WO2021236998) or provisional patent applications (63/271,627) filed by Columbia University for a number of SARS-CoV-2 neutralizing antibodies described in this manuscript. Both sets of applications are under review. D.D.H. is a co-founder of TaiMed Biologics and RenBio, consultant to WuXi Biologics and Brii Biosciences, and board director for Vicarious Surgical.
Figures


Similar articles
-
Susceptibility of SARS-CoV-2 Omicron Variants to Therapeutic Monoclonal Antibodies: Systematic Review and Meta-analysis.Microbiol Spectr. 2022 Aug 31;10(4):e0092622. doi: 10.1128/spectrum.00926-22. Epub 2022 Jun 14. Microbiol Spectr. 2022. PMID: 35700134 Free PMC article.
-
SARS-CoV-2 Omicron sublineages exhibit distinct antibody escape patterns.Cell Host Microbe. 2022 Sep 14;30(9):1231-1241.e6. doi: 10.1016/j.chom.2022.07.002. Epub 2022 Jul 7. Cell Host Microbe. 2022. PMID: 35921836 Free PMC article.
-
The Omicron variant of concern: Diversification and convergent evolution in spike protein, and escape from anti-Spike monoclonal antibodies.Drug Resist Updat. 2022 Dec;65:100882. doi: 10.1016/j.drup.2022.100882. Epub 2022 Oct 3. Drug Resist Updat. 2022. PMID: 36260961 Free PMC article. Review.
-
Resistance of SARS-CoV-2 Omicron BA.1 and BA.2 Variants to Vaccine-Elicited Sera and Therapeutic Monoclonal Antibodies.Viruses. 2022 Jun 18;14(6):1334. doi: 10.3390/v14061334. Viruses. 2022. PMID: 35746806 Free PMC article.
-
Evolution of the SARS-CoV-2 omicron variants BA.1 to BA.5: Implications for immune escape and transmission.Rev Med Virol. 2022 Sep;32(5):e2381. doi: 10.1002/rmv.2381. Epub 2022 Jul 20. Rev Med Virol. 2022. PMID: 35856385 Free PMC article. Review.
Cited by
-
The Rise and Fall of SARS-CoV-2 Variants and Ongoing Diversification of Omicron.Viruses. 2022 Sep 10;14(9):2009. doi: 10.3390/v14092009. Viruses. 2022. PMID: 36146815 Free PMC article. Review.
-
Outpatient Management of COVID-19: A Primer for the Dermatologist.Curr Dermatol Rep. 2022;11(4):318-327. doi: 10.1007/s13671-022-00368-3. Epub 2022 Aug 20. Curr Dermatol Rep. 2022. PMID: 36035078 Free PMC article. Review.
-
Short-course early outpatient remdesivir prevents severe disease due to COVID-19 in organ transplant recipients during the omicron BA.2 wave.Am J Transplant. 2023 Jan;23(1):78-83. doi: 10.1111/ajt.17199. Epub 2023 Jan 11. Am J Transplant. 2023. PMID: 36148607 Free PMC article.
-
Antibody-mediated immunity to SARS-CoV-2 spike.Adv Immunol. 2022;154:1-69. doi: 10.1016/bs.ai.2022.07.001. Epub 2022 Aug 22. Adv Immunol. 2022. PMID: 36038194 Free PMC article. Review.
-
Does early combination vs. Monotherapy improve clinical outcomes of clinically extremely vulnerable patients with COVID-19? Results from a retrospective propensity-weighted analysis.Eur J Med Res. 2024 Oct 4;29(1):484. doi: 10.1186/s40001-024-02062-5. Eur J Med Res. 2024. PMID: 39367485 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

