Large-scale survey and database of high affinity ligands for peptide recognition modules
- PMID: 33438817
- PMCID: PMC7724964
- DOI: 10.15252/msb.20199310
Large-scale survey and database of high affinity ligands for peptide recognition modules
Abstract
Many proteins involved in signal transduction contain peptide recognition modules (PRMs) that recognize short linear motifs (SLiMs) within their interaction partners. Here, we used large-scale peptide-phage display methods to derive optimal ligands for 163 unique PRMs representing 79 distinct structural families. We combined the new data with previous data that we collected for the large SH3, PDZ, and WW domain families to assemble a database containing 7,984 unique peptide ligands for 500 PRMs representing 82 structural families. For 74 PRMs, we acquired enough new data to map the specificity profiles in detail and derived position weight matrices and binding specificity logos based on multiple peptide ligands. These analyses showed that optimal peptide ligands resembled peptides observed in existing structures of PRM-ligand complexes, indicating that a large majority of the phage-derived peptides are likely to target natural peptide-binding sites and could thus act as inhibitors of natural protein-protein interactions. The complete dataset has been assembled in an online database (http://www.prm-db.org) that will enable many structural, functional, and biological studies of PRMs and SLiMs.
Keywords: domain specificity; peptide inhibitors; peptide library; peptide recognition modules; phage display.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
Distribution of specificity profile lengths.
Distribution of total specificity potential (SPt) scores.
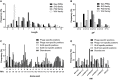
Frequencies of amino acids at specific and non‐specific positions in phage‐derived peptides and SLiMs, and in disorderome peptides. Amino acids are ordered from highest to lowest hydrophobicity measured by Roseman’s Hydropathy Index (RHI), which is shown below each amino acid denoted by the single‐letter code.
Distribution of the RHI for specific and non‐specific positions in phage‐derived peptides and SLiMs and in disorderome peptides.
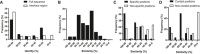
Distribution of sequence identities between the studied PRMs and the matched PRM structures for the full sequence or the sequence of the peptide‐binding interface region.
Distribution of sequence similarities between the ligand in the complex structure and the most similar phage‐derived peptide for all 135 PRMs.
Distribution of sequence similarities between the ligand in the complex structure and the most similar phage‐derived peptide for the 63 PRMs with specificity profiles (Fig 1), either at specific positions or non‐specific positions.
Distribution of sequence similarities between the ligand in the complex structure and the most similar phage‐derived peptide at contact and non‐contact positions in the structure.


- A–C
Depicted are the 17 PRMs for which the ligand in the structure contains (A) pSer/pThr, (B) pTyr, or (C) meArg/meLys. The name of the protein from which the studied PRM was derived is listed at the top with the PRM family name in parenthesis, followed by the specificity profile determined from phage‐derived peptides, if available. Below, the alignment shows the sequences of a phage‐derived peptide (top) and the peptide ligand from the best‐matched PRM‐ligand complex in the PDB (bottom). Similar residues in the two peptides are shaded gray and residues that make contact with the PRM in the structure are underlined. Filled circles below the alignment indicate residues that contain PTMs. The structure of the best‐matched PRM‐ligand complex from the PDB is shown with the PRM and peptide ligand main chains rendered as gray or green ribbons, respectively. Red and blue spheres denote ligand positions that are similar to the phage‐derived peptide and are contact or non‐contact positions, respectively. Side chains that contain PTMs are shown as yellow sticks. The peptide main chain is only depicted for those residues that are shown in the alignment with the phage‐derived peptide. The PDB entry code is shown above each structure.

Similar articles
-
Domain Interaction Footprint: a multi-classification approach to predict domain-peptide interactions.Bioinformatics. 2009 Jul 1;25(13):1632-9. doi: 10.1093/bioinformatics/btp264. Epub 2009 Apr 17. Bioinformatics. 2009. PMID: 19376827
-
Studying binding specificities of peptide recognition modules by high-throughput phage display selections.Methods Mol Biol. 2011;781:87-97. doi: 10.1007/978-1-61779-276-2_6. Methods Mol Biol. 2011. PMID: 21877279
-
Identifying specificity profiles for peptide recognition modules from phage-displayed peptide libraries.Nat Protoc. 2007;2(6):1368-86. doi: 10.1038/nprot.2007.151. Nat Protoc. 2007. PMID: 17545975
-
Functional genomics of intracellular peptide recognition domains with combinatorial biology methods.Curr Opin Chem Biol. 2003 Feb;7(1):97-102. doi: 10.1016/s1367-5931(02)00011-x. Curr Opin Chem Biol. 2003. PMID: 12547433 Review.
-
Designing scaffolds of peptides for phage display libraries.J Biosci Bioeng. 2005 May;99(5):448-56. doi: 10.1263/jbb.99.448. J Biosci Bioeng. 2005. PMID: 16233816 Review.
Cited by
-
Peptide-binding specificity prediction using fine-tuned protein structure prediction networks.Proc Natl Acad Sci U S A. 2023 Feb 28;120(9):e2216697120. doi: 10.1073/pnas.2216697120. Epub 2023 Feb 21. Proc Natl Acad Sci U S A. 2023. PMID: 36802421 Free PMC article.
-
Large scale discovery of coronavirus-host factor protein interaction motifs reveals SARS-CoV-2 specific mechanisms and vulnerabilities.Nat Commun. 2021 Nov 19;12(1):6761. doi: 10.1038/s41467-021-26498-z. Nat Commun. 2021. PMID: 34799561 Free PMC article.
-
Discovery of an exosite on the SOCS2-SH2 domain that enhances SH2 binding to phosphorylated ligands.Nat Commun. 2021 Dec 2;12(1):7032. doi: 10.1038/s41467-021-26983-5. Nat Commun. 2021. PMID: 34857742 Free PMC article.
-
Immunopeptides: immunomodulatory strategies and prospects for ocular immunity applications.Front Immunol. 2024 Jul 15;15:1406762. doi: 10.3389/fimmu.2024.1406762. eCollection 2024. Front Immunol. 2024. PMID: 39076973 Free PMC article. Review.
-
Large-scale phage-based screening reveals extensive pan-viral mimicry of host short linear motifs.Nat Commun. 2023 Apr 26;14(1):2409. doi: 10.1038/s41467-023-38015-5. Nat Commun. 2023. PMID: 37100772 Free PMC article.
References
-
- Betts MJ, Russell RB (2003) Amino acid properties and consequences of substitutions Bioinformatics for geneticists, pp 289–316. John Wiley & Sons, Ltd;
-
- Carducci M, Perfetto L, Briganti L, Paoluzi S, Costa S, Zerweck J, Schutkowski M, Castagnoli L, Cesareni G (2012) The protein interaction network mediated by human SH3 domains. Biotechnol Adv 30: 4–15 - PubMed
-
- Chatr‐aryamontri Andrew, Oughtred Rose, Boucher Lorrie, Rust Jennifer, Chang Christie, Kolas Nadine K, O'Donnell Lara, Oster Sara, Theesfeld Chandra, Sellam Adnane, Stark Chris, Breitkreutz Bobby‐Joe, Dolinski Kara, Tyers Mike (2017) The BioGRID interaction database: 2017 update. Nucleic Acids Res 45: D369–D379 - PMC - PubMed
-
- Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ (2012) Attributes of short linear motifs. Mol BioSyst 8: 268–281 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

