Progenitor identification and SARS-CoV-2 infection in human distal lung organoids
- PMID: 33238290
- PMCID: PMC8003326
- DOI: 10.1038/s41586-020-3014-1
Progenitor identification and SARS-CoV-2 infection in human distal lung organoids
Abstract
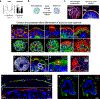
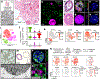
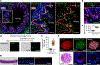
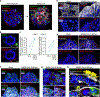
The distal lung contains terminal bronchioles and alveoli that facilitate gas exchange. Three-dimensional in vitro human distal lung culture systems would strongly facilitate the investigation of pathologies such as interstitial lung disease, cancer and coronavirus disease 2019 (COVID-19) pneumonia caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Here we describe the development of a long-term feeder-free, chemically defined culture system for distal lung progenitors as organoids derived from single adult human alveolar epithelial type II (AT2) or KRT5+ basal cells. AT2 organoids were able to differentiate into AT1 cells, and basal cell organoids developed lumens lined with differentiated club and ciliated cells. Single-cell analysis of KRT5+ cells in basal organoids revealed a distinct population of ITGA6+ITGB4+ mitotic cells, whose offspring further segregated into a TNFRSF12Ahi subfraction that comprised about ten per cent of KRT5+ basal cells. This subpopulation formed clusters within terminal bronchioles and exhibited enriched clonogenic organoid growth activity. We created distal lung organoids with apical-out polarity to present ACE2 on the exposed external surface, facilitating infection of AT2 and basal cultures with SARS-CoV-2 and identifying club cells as a target population. This long-term, feeder-free culture of human distal lung organoids, coupled with single-cell analysis, identifies functional heterogeneity among basal cells and establishes a facile in vitro organoid model of human distal lung infections, including COVID-19-associated pneumonia.
Conflict of interest statement
Figures






Update of
-
Progenitor identification and SARS-CoV-2 infection in long-term human distal lung organoid cultures.bioRxiv [Preprint]. 2020 Jul 27:2020.07.27.212076. doi: 10.1101/2020.07.27.212076. bioRxiv. 2020. Update in: Nature. 2020 Dec;588(7839):670-675. doi: 10.1038/s41586-020-3014-1. PMID: 32743583 Free PMC article. Updated. Preprint.
Similar articles
-
Progenitor identification and SARS-CoV-2 infection in long-term human distal lung organoid cultures.bioRxiv [Preprint]. 2020 Jul 27:2020.07.27.212076. doi: 10.1101/2020.07.27.212076. bioRxiv. 2020. Update in: Nature. 2020 Dec;588(7839):670-675. doi: 10.1038/s41586-020-3014-1. PMID: 32743583 Free PMC article. Updated. Preprint.
-
Host metabolism dysregulation and cell tropism identification in human airway and alveolar organoids upon SARS-CoV-2 infection.Protein Cell. 2021 Sep;12(9):717-733. doi: 10.1007/s13238-020-00811-w. Epub 2020 Dec 12. Protein Cell. 2021. PMID: 33314005 Free PMC article.
-
An organoid-derived bronchioalveolar model for SARS-CoV-2 infection of human alveolar type II-like cells.EMBO J. 2021 Mar 1;40(5):e105912. doi: 10.15252/embj.2020105912. Epub 2021 Jan 11. EMBO J. 2021. PMID: 33283287 Free PMC article.
-
SARS-CoV-2 Infection and Disease Modelling Using Stem Cell Technology and Organoids.Int J Mol Sci. 2021 Feb 26;22(5):2356. doi: 10.3390/ijms22052356. Int J Mol Sci. 2021. PMID: 33652988 Free PMC article. Review.
-
Basal-like Progenitor Cells: A Review of Dysplastic Alveolar Regeneration and Remodeling in Lung Repair.Stem Cell Reports. 2020 Nov 10;15(5):1015-1025. doi: 10.1016/j.stemcr.2020.09.006. Epub 2020 Oct 15. Stem Cell Reports. 2020. PMID: 33065046 Free PMC article. Review.
Cited by
-
Alveolar type 2 cells marker gene SFTPC inhibits epithelial-to-mesenchymal transition by upregulating SOX7 and suppressing WNT/β-catenin pathway in non-small cell lung cancer.Front Oncol. 2024 Sep 13;14:1448379. doi: 10.3389/fonc.2024.1448379. eCollection 2024. Front Oncol. 2024. PMID: 39346732 Free PMC article.
-
Identification of a Novel Subset of Human Airway Epithelial Basal Stem Cells.Int J Mol Sci. 2024 Sep 12;25(18):9863. doi: 10.3390/ijms25189863. Int J Mol Sci. 2024. PMID: 39337350 Free PMC article.
-
Chronic lung inflammation and CK14+ basal cell proliferation induce persistent alveolar-bronchiolization in SARS-CoV-2-infected hamsters.EBioMedicine. 2024 Oct;108:105363. doi: 10.1016/j.ebiom.2024.105363. Epub 2024 Sep 25. EBioMedicine. 2024. PMID: 39326207 Free PMC article.
-
Organoids: development and applications in disease models, drug discovery, precision medicine, and regenerative medicine.MedComm (2020). 2024 Sep 21;5(10):e735. doi: 10.1002/mco2.735. eCollection 2024 Oct. MedComm (2020). 2024. PMID: 39309690 Free PMC article. Review.
-
Advances in an In Vitro Tuberculosis Infection Model Using Human Lung Organoids for Host-Directed Therapies.PLoS Pathog. 2024 Jul 25;20(7):e1012295. doi: 10.1371/journal.ppat.1012295. eCollection 2024 Jul. PLoS Pathog. 2024. PMID: 39052544 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- T32 GM007365/GM/NIGMS NIH HHS/United States
- R01 AI157155/AI/NIAID NIH HHS/United States
- DK11572802/NH/NIH HHS/United States
- U24 DK085532/DK/NIDDK NIH HHS/United States
- U01 DE025188/DE/NIDCR NIH HHS/United States
- UH3 CA255135/CA/NCI NIH HHS/United States
- U01 DK085532/DK/NIDDK NIH HHS/United States
- UG3 HL145623/HL/NHLBI NIH HHS/United States
- T32 AI007502-23/NH/NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- T32 AI007502/AI/NIAID NIH HHS/United States
- R01 HL142549/HL/NHLBI NIH HHS/United States
- T32 GM007365-44/NH/NIH HHS/United States
- U01 CA176299/CA/NCI NIH HHS/United States
- R56 AI111460/AI/NIAID NIH HHS/United States
- 5R01HL14254902/NH/NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- K08 DE027730/DE/NIDCR NIH HHS/United States
- U01 DK085527/DK/NIDDK NIH HHS/United States
- U01 CA217851/CA/NCI NIH HHS/United States
- U19 AI116484/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

