In vivo CRISPR screening for phenotypic targets of the mir-35-42 family in C. elegans
- PMID: 32820039
- PMCID: PMC7462058
- DOI: 10.1101/gad.339333.120
In vivo CRISPR screening for phenotypic targets of the mir-35-42 family in C. elegans
Erratum in
-
Corrigendum: In vivo CRISPR screening for phenotypic targets of the mir-35-42 family in C. elegans.Genes Dev. 2021 Dec 1;35(23-24):1693. doi: 10.1101/gad.349209.121. Genes Dev. 2021. PMID: 34862181 Free PMC article. No abstract available.
Abstract
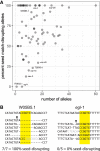
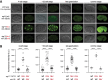
Identifying miRNA target genes is difficult, and delineating which targets are the most biologically important is even more difficult. We devised a novel strategy to test the phenotypic impact of individual microRNA-target interactions by disrupting each predicted miRNA-binding site by CRISPR-Cas9 genome editing in C. elegans We developed a multiplexed negative selection screening approach in which edited loci are deep sequenced, and candidate sites are prioritized based on apparent selection pressure against mutations that disrupt miRNA binding. Importantly, our screen was conducted in vivo on mutant animals, allowing us to interrogate organism-level phenotypes. We used this approach to screen for phenotypic targets of the essential mir-35-42 family. By generating 1130 novel 3'UTR alleles across all predicted targets, we identified egl-1 as a phenotypic target whose derepression partially phenocopies the mir-35-42 mutant phenotype by inducing embryonic lethality and low fecundity. These phenotypes can be rescued by compensatory CRISPR mutations that retarget mir-35 to the mutant egl-1 3'UTR. This study demonstrates that the application of in vivo whole organismal CRISPR screening has great potential to accelerate the discovery of phenotypic negative regulatory elements in the noncoding genome.
Keywords: C. elegans; CRISPR; in vivo screening; miRNA targets; miRNAs; mutational profiling; negative selection screening.
© 2020 Yang et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


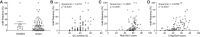
Similar articles
-
Parallel genetics of regulatory sequences using scalable genome editing in vivo.Cell Rep. 2021 Apr 13;35(2):108988. doi: 10.1016/j.celrep.2021.108988. Cell Rep. 2021. PMID: 33852857
-
Staufen Negatively Modulates MicroRNA Activity in Caenorhabditis elegans.G3 (Bethesda). 2016 May 3;6(5):1227-37. doi: 10.1534/g3.116.027300. G3 (Bethesda). 2016. PMID: 26921297 Free PMC article.
-
miR-124/ATF-6, a novel lifespan extension pathway of Astragalus polysaccharide in Caenorhabditis elegans.J Cell Biochem. 2015 Feb;116(2):242-51. doi: 10.1002/jcb.24961. J Cell Biochem. 2015. PMID: 25186652
-
The application of somatic CRISPR-Cas9 to conditional genome editing in Caenorhabditis elegans.Genesis. 2016 Apr;54(4):170-81. doi: 10.1002/dvg.22932. Epub 2016 Apr 14. Genesis. 2016. PMID: 26934570 Review.
-
[CRISPR-Cas9 mediated genome editing in Caenorhabditis elegans].Sheng Wu Gong Cheng Xue Bao. 2017 Oct 25;33(10):1693-1699. doi: 10.13345/j.cjb.170177. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 29082717 Review. Chinese.
Cited by
-
A new perspective on microRNA-guided gene regulation specificity, and its potential generalization to transcription factors and RNA-binding proteins.Nucleic Acids Res. 2024 Sep 9;52(16):9360-9368. doi: 10.1093/nar/gkae694. Nucleic Acids Res. 2024. PMID: 39149906 Free PMC article. Review.
-
Catalytic residues of microRNA Argonautes play a modest role in microRNA star strand destabilization in C. elegans.Nucleic Acids Res. 2024 May 22;52(9):4985-5001. doi: 10.1093/nar/gkae170. Nucleic Acids Res. 2024. PMID: 38471816 Free PMC article.
-
Looking for a needle in a haystack: de novo phenotypic target identification reveals Hippo pathway-mediated miR-202 regulation of egg production.Nucleic Acids Res. 2024 Jan 25;52(2):738-754. doi: 10.1093/nar/gkad1154. Nucleic Acids Res. 2024. PMID: 38059397 Free PMC article.
-
The miRNA-target interactions: An underestimated intricacy.Nucleic Acids Res. 2024 Feb 28;52(4):1544-1557. doi: 10.1093/nar/gkad1142. Nucleic Acids Res. 2024. PMID: 38033323 Free PMC article. Review.
-
microRNAs in action: biogenesis, function and regulation.Nat Rev Genet. 2023 Dec;24(12):816-833. doi: 10.1038/s41576-023-00611-y. Epub 2023 Jun 28. Nat Rev Genet. 2023. PMID: 37380761 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
