Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses
- PMID: 32094589
- PMCID: PMC7095430
- DOI: 10.1038/s41564-020-0688-y
Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses
Abstract
Over the past 20 years, several coronaviruses have crossed the species barrier into humans, causing outbreaks of severe, and often fatal, respiratory illness. Since SARS-CoV was first identified in animal markets, global viromics projects have discovered thousands of coronavirus sequences in diverse animals and geographic regions. Unfortunately, there are few tools available to functionally test these viruses for their ability to infect humans, which has severely hampered efforts to predict the next zoonotic viral outbreak. Here, we developed an approach to rapidly screen lineage B betacoronaviruses, such as SARS-CoV and the recent SARS-CoV-2, for receptor usage and their ability to infect cell types from different species. We show that host protease processing during viral entry is a significant barrier for several lineage B viruses and that bypassing this barrier allows several lineage B viruses to enter human cells through an unknown receptor. We also demonstrate how different lineage B viruses can recombine to gain entry into human cells, and confirm that human ACE2 is the receptor for the recently emerging SARS-CoV-2.
Conflict of interest statement
The authors declare no competing interests.
Figures

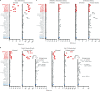

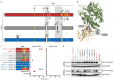



Update of
-
Functional assessment of cell entry and receptor usage for lineage B β-coronaviruses, including 2019-nCoV.bioRxiv [Preprint]. 2020 Jan 22:2020.01.22.915660. doi: 10.1101/2020.01.22.915660. bioRxiv. 2020. Update in: Nat Microbiol. 2020 Apr;5(4):562-569. doi: 10.1038/s41564-020-0688-y. PMID: 32511294 Free PMC article. Updated. Preprint.
Comment in
-
Functionally assessing coronavirus entry.Nat Med. 2020 Apr;26(4):465. doi: 10.1038/s41591-020-0847-y. Nat Med. 2020. PMID: 32273603 No abstract available.
Similar articles
-
Receptor Recognition by the Novel Coronavirus from Wuhan: an Analysis Based on Decade-Long Structural Studies of SARS Coronavirus.J Virol. 2020 Mar 17;94(7):e00127-20. doi: 10.1128/JVI.00127-20. Print 2020 Mar 17. J Virol. 2020. PMID: 31996437 Free PMC article.
-
Broad and Differential Animal Angiotensin-Converting Enzyme 2 Receptor Usage by SARS-CoV-2.J Virol. 2020 Aug 31;94(18):e00940-20. doi: 10.1128/JVI.00940-20. Print 2020 Aug 31. J Virol. 2020. PMID: 32661139 Free PMC article.
-
Composition and divergence of coronavirus spike proteins and host ACE2 receptors predict potential intermediate hosts of SARS-CoV-2.J Med Virol. 2020 Jun;92(6):595-601. doi: 10.1002/jmv.25726. Epub 2020 Mar 11. J Med Virol. 2020. PMID: 32100877 Free PMC article.
-
SARS-CoV-2 pandemic and research gaps: Understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy.Theranostics. 2020 Jun 12;10(16):7448-7464. doi: 10.7150/thno.48076. eCollection 2020. Theranostics. 2020. PMID: 32642005 Free PMC article. Review.
-
Angiotensin-converting enzyme 2: The old door for new severe acute respiratory syndrome coronavirus 2 infection.Rev Med Virol. 2020 Sep;30(5):e2122. doi: 10.1002/rmv.2122. Epub 2020 Jun 30. Rev Med Virol. 2020. PMID: 32602627 Free PMC article. Review.
Cited by
-
Human-Based Advanced in vitro Approaches to Investigate Lung Fibrosis and Pulmonary Effects of COVID-19.Front Med (Lausanne). 2021 May 7;8:644678. doi: 10.3389/fmed.2021.644678. eCollection 2021. Front Med (Lausanne). 2021. PMID: 34026781 Free PMC article. Review.
-
SARS-CoV-2 infections elicit higher levels of original antigenic sin antibodies compared with SARS-CoV-2 mRNA vaccinations.Cell Rep. 2022 Oct 18;41(3):111496. doi: 10.1016/j.celrep.2022.111496. Cell Rep. 2022. PMID: 36261003 Free PMC article.
-
Production and secretion of functional SARS-CoV-2 spike protein in Chlamydomonas reinhardtii.Front Plant Sci. 2022 Sep 20;13:988870. doi: 10.3389/fpls.2022.988870. eCollection 2022. Front Plant Sci. 2022. PMID: 36204065 Free PMC article.
-
An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates.Sci Transl Med. 2020 Aug 5;12(555):eabc9396. doi: 10.1126/scitranslmed.abc9396. Epub 2020 Jul 20. Sci Transl Med. 2020. PMID: 32690628 Free PMC article.
-
GB-2 inhibits ACE2 and TMPRSS2 expression: In vivo and in vitro studies.Biomed Pharmacother. 2020 Dec;132:110816. doi: 10.1016/j.biopha.2020.110816. Epub 2020 Oct 10. Biomed Pharmacother. 2020. PMID: 33049583 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

