Structural basis for human coronavirus attachment to sialic acid receptors
- PMID: 31160783
- PMCID: PMC6554059
- DOI: 10.1038/s41594-019-0233-y
Structural basis for human coronavirus attachment to sialic acid receptors
Abstract
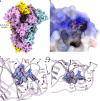
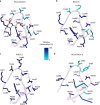
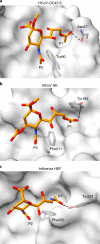
Coronaviruses cause respiratory tract infections in humans and outbreaks of deadly pneumonia worldwide. Infections are initiated by the transmembrane spike (S) glycoprotein, which binds to host receptors and fuses the viral and cellular membranes. To understand the molecular basis of coronavirus attachment to oligosaccharide receptors, we determined cryo-EM structures of coronavirus OC43 S glycoprotein trimer in isolation and in complex with a 9-O-acetylated sialic acid. We show that the ligand binds with fast kinetics to a surface-exposed groove and that interactions at the identified site are essential for S-mediated viral entry into host cells, but free monosaccharide does not trigger fusogenic conformational changes. The receptor-interacting site is conserved in all coronavirus S glycoproteins that engage 9-O-acetyl-sialogycans, with an architecture similar to those of the ligand-binding pockets of coronavirus hemagglutinin esterases and influenza virus C/D hemagglutinin-esterase fusion glycoproteins. Our results demonstrate these viruses evolved similar strategies to engage sialoglycans at the surface of target cells.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Human coronaviruses OC43 and HKU1 bind to 9-O-acetylated sialic acids via a conserved receptor-binding site in spike protein domain A.Proc Natl Acad Sci U S A. 2019 Feb 12;116(7):2681-2690. doi: 10.1073/pnas.1809667116. Epub 2019 Jan 24. Proc Natl Acad Sci U S A. 2019. PMID: 30679277 Free PMC article.
-
Human Coronavirus HKU1 Spike Protein Uses O-Acetylated Sialic Acid as an Attachment Receptor Determinant and Employs Hemagglutinin-Esterase Protein as a Receptor-Destroying Enzyme.J Virol. 2015 Jul;89(14):7202-13. doi: 10.1128/JVI.00854-15. Epub 2015 Apr 29. J Virol. 2015. PMID: 25926653 Free PMC article.
-
Coronavirus hemagglutinin-esterase and spike proteins coevolve for functional balance and optimal virion avidity.Proc Natl Acad Sci U S A. 2020 Oct 13;117(41):25759-25770. doi: 10.1073/pnas.2006299117. Epub 2020 Sep 29. Proc Natl Acad Sci U S A. 2020. PMID: 32994342 Free PMC article.
-
SARS-CoV-2 Evolutionary Adaptation toward Host Entry and Recognition of Receptor O-Acetyl Sialylation in Virus-Host Interaction.Int J Mol Sci. 2020 Jun 26;21(12):4549. doi: 10.3390/ijms21124549. Int J Mol Sci. 2020. PMID: 32604730 Free PMC article. Review.
-
Structure, Function, and Evolution of Coronavirus Spike Proteins.Annu Rev Virol. 2016 Sep 29;3(1):237-261. doi: 10.1146/annurev-virology-110615-042301. Epub 2016 Aug 25. Annu Rev Virol. 2016. PMID: 27578435 Free PMC article. Review.
Cited by
-
Recombinant human plasma gelsolin reverses increased permeability of the blood-brain barrier induced by the spike protein of the SARS-CoV-2 virus.J Neuroinflammation. 2022 Nov 24;19(1):282. doi: 10.1186/s12974-022-02642-4. J Neuroinflammation. 2022. PMID: 36434734 Free PMC article.
-
Intra-species sialic acid polymorphism in humans: a common niche for influenza and coronavirus pandemics?Emerg Microbes Infect. 2021 Dec;10(1):1191-1199. doi: 10.1080/22221751.2021.1935329. Emerg Microbes Infect. 2021. PMID: 34049471 Free PMC article. Review.
-
Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies.Cell. 2020 Aug 20;182(4):828-842.e16. doi: 10.1016/j.cell.2020.06.025. Epub 2020 Jun 24. Cell. 2020. PMID: 32645326 Free PMC article.
-
Targeting Viral ORF3a Protein: A New Approach to Mitigate COVID-19 Induced Immune Cell Apoptosis and Associated Respiratory Complications.Adv Pharm Bull. 2023 Nov;13(4):678-687. doi: 10.34172/apb.2023.069. Epub 2023 Jan 23. Adv Pharm Bull. 2023. PMID: 38022818 Free PMC article. Review.
-
Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19.Acta Pharmacol Sin. 2020 Sep;41(9):1141-1149. doi: 10.1038/s41401-020-0485-4. Epub 2020 Aug 3. Acta Pharmacol Sin. 2020. PMID: 32747721 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

