Biochemical and structural investigations clarify the substrate selectivity of the 2-oxoglutarate oxygenase JMJD6
- PMID: 31147442
- PMCID: PMC6663879
- DOI: 10.1074/jbc.RA119.008693
Biochemical and structural investigations clarify the substrate selectivity of the 2-oxoglutarate oxygenase JMJD6
Abstract
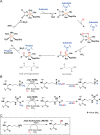
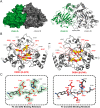
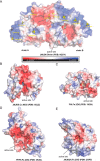
JmjC domain-containing protein 6 (JMJD6) is a 2-oxoglutarate (2OG)-dependent oxygenase linked to various cellular processes, including splicing regulation, histone modification, transcriptional pause release, hypoxia sensing, and cancer. JMJD6 is reported to catalyze hydroxylation of lysine residue(s) of histones, the tumor-suppressor protein p53, and splicing regulatory proteins, including u2 small nuclear ribonucleoprotein auxiliary factor 65-kDa subunit (U2AF65). JMJD6 is also reported to catalyze N-demethylation of N-methylated (both mono- and di-methylated) arginine residues of histones and other proteins, including HSP70 (heat-shock protein 70), estrogen receptor α, and RNA helicase A. Here, we report MS- and NMR-based kinetic assays employing purified JMJD6 and multiple substrate fragment sequences, the results of which support the assignment of purified JMJD6 as a lysyl hydroxylase. By contrast, we did not observe N-methyl arginyl N-demethylation with purified JMJD6. Biophysical analyses, including crystallographic analyses of JMJD6Δ344-403 in complex with iron and 2OG, supported its assignment as a lysyl hydroxylase rather than an N-methyl arginyl-demethylase. The screening results supported some, but not all, of the assigned JMJD6 substrates and identified other potential JMJD6 substrates. We envision these results will be useful in cellular and biological work on the substrates and functions of JMJD6 and in the development of selective inhibitors of human 2OG oxygenases.
Keywords: 2-oxoglutarate and iron dependent dioxygenase; C-5 hydroxylysine; JMJD6; JmjC domain-containing protein 6; RNA splicing; X-ray crystallography; dioxygenase; enzyme catalysis; enzyme structure; hydroxylase; hydroxylysine (Hyl); hypoxia; metalloenzyme; substrate specificity.
© 2019 Islam et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




Similar articles
-
The oxygenase Jmjd6--a case study in conflicting assignments.Biochem J. 2015 Jun 1;468(2):191-202. doi: 10.1042/BJ20150278. Biochem J. 2015. PMID: 25997831 Review.
-
Studies on the catalytic domains of multiple JmjC oxygenases using peptide substrates.Epigenetics. 2014 Dec;9(12):1596-603. doi: 10.4161/15592294.2014.983381. Epigenetics. 2014. PMID: 25625844 Free PMC article.
-
The hydroxylation activity of Jmjd6 is required for its homo-oligomerization.J Cell Biochem. 2012 May;113(5):1663-70. doi: 10.1002/jcb.24035. J Cell Biochem. 2012. PMID: 22189873
-
Widespread hydroxylation of unstructured lysine-rich protein domains by JMJD6.Proc Natl Acad Sci U S A. 2022 Aug 9;119(32):e2201483119. doi: 10.1073/pnas.2201483119. Epub 2022 Aug 5. Proc Natl Acad Sci U S A. 2022. PMID: 35930668 Free PMC article.
-
Jmjd6, a JmjC Dioxygenase with Many Interaction Partners and Pleiotropic Functions.Front Genet. 2017 Mar 16;8:32. doi: 10.3389/fgene.2017.00032. eCollection 2017. Front Genet. 2017. PMID: 28360925 Free PMC article. Review.
Cited by
-
To Erase or Not to Erase: Non-Canonical Catalytic Functions and Non-Catalytic Functions of Members of Histone Lysine Demethylase Families.Int J Mol Sci. 2024 Jun 24;25(13):6900. doi: 10.3390/ijms25136900. Int J Mol Sci. 2024. PMID: 39000010 Free PMC article. Review.
-
JMJD6 Is a Druggable Oxygenase That Regulates AR-V7 Expression in Prostate Cancer.Cancer Res. 2021 Feb 15;81(4):1087-1100. doi: 10.1158/0008-5472.CAN-20-1807. Cancer Res. 2021. PMID: 33822745 Free PMC article.
-
The Ligand of Ate1 is intrinsically disordered and participates in nucleolar phase separation regulated by Jumonji Domain Containing 6.Proc Natl Acad Sci U S A. 2021 Jan 5;118(1):e2015887118. doi: 10.1073/pnas.2015887118. Epub 2020 Dec 21. Proc Natl Acad Sci U S A. 2021. PMID: 33443146 Free PMC article.
-
Promotion of adipogenesis by JMJD6 requires the AT hook-like domain and is independent of its catalytic function.PLoS One. 2019 Aug 20;14(8):e0216015. doi: 10.1371/journal.pone.0216015. eCollection 2019. PLoS One. 2019. PMID: 31430278 Free PMC article.
-
Protein arginine methylation: from enigmatic functions to therapeutic targeting.Nat Rev Drug Discov. 2021 Jul;20(7):509-530. doi: 10.1038/s41573-021-00159-8. Epub 2021 Mar 19. Nat Rev Drug Discov. 2021. PMID: 33742187 Review.
References
-
- Schofield C. J., and Hausinger R. P. (2015) in 2-Oxoglutarate-dependent Oxygenases, pp. 1–58, The Royal Society of Chemistry, Cambridge, UK
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

