WRN helicase is a synthetic lethal target in microsatellite unstable cancers
- PMID: 30971823
- PMCID: PMC6580861
- DOI: 10.1038/s41586-019-1102-x
WRN helicase is a synthetic lethal target in microsatellite unstable cancers
Abstract
Synthetic lethality-an interaction between two genetic events through which the co-occurrence of these two genetic events leads to cell death, but each event alone does not-can be exploited for cancer therapeutics1. DNA repair processes represent attractive synthetic lethal targets, because many cancers exhibit an impairment of a DNA repair pathway, which can lead to dependence on specific repair proteins2. The success of poly(ADP-ribose) polymerase 1 (PARP-1) inhibitors in cancers with deficiencies in homologous recombination highlights the potential of this approach3. Hypothesizing that other DNA repair defects would give rise to synthetic lethal relationships, we queried dependencies in cancers with microsatellite instability (MSI), which results from deficient DNA mismatch repair. Here we analysed data from large-scale silencing screens using CRISPR-Cas9-mediated knockout and RNA interference, and found that the RecQ DNA helicase WRN was selectively essential in MSI models in vitro and in vivo, yet dispensable in models of cancers that are microsatellite stable. Depletion of WRN induced double-stranded DNA breaks and promoted apoptosis and cell cycle arrest selectively in MSI models. MSI cancer models required the helicase activity of WRN, but not its exonuclease activity. These findings show that WRN is a synthetic lethal vulnerability and promising drug target for MSI cancers.
Conflict of interest statement
Competing Interest
A.J.B. receives research funding from Merck and Novartis. D.E.R. receives research funding from the Functional Genomics Consortium (Abbvie, Jannsen, Merck, Vir) and is a director of Addgene. A.T. consults for Tango Therapeutics. T.R.G. has advised Foundation Medicine, Glaxo SmithKline, Sherlock Biosciences, Forma Therapeutics. A.D.A. consults for Lilly Oncology, EMD Serono, Intellia Therapeutics, Sierra Oncology, Formation Biologics, Cyteir Therapeutics, consults and holds stock in Ideaya, and co-founded and holds stock in Cedilla Therapeutics. G.G. receives research funding from IBM and Pharmacyclics and is an inventor on multiple patent applications related to bioinformatic tools, including applications related to MuTect, ABSOLUTE, MSMuTect, MSMutSig and MSIClass. Y.E.M. is an inventor on patent applications related to the bioinformatic tools, MSMuTect, MSMutSig and MSIClass. The Broad Institute filed a US patent application related to the target described in this manuscript.
Figures







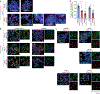
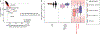
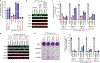

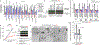
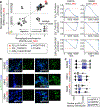
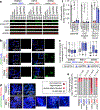
Comment in
-
Lethal clues to cancer-cell vulnerability.Nature. 2019 Apr;568(7753):463-464. doi: 10.1038/d41586-019-01086-w. Nature. 2019. PMID: 31000834 No abstract available.
-
Prioritizing synthetic lethal targets with functional genomics.Nat Rev Cancer. 2019 Jun;19(6):305. doi: 10.1038/s41568-019-0147-3. Nat Rev Cancer. 2019. PMID: 31036884 No abstract available.
-
CRISPR Screens Single Out WRN.Cancer Discov. 2019 Jul;9(7):OF6. doi: 10.1158/2159-8290.CD-NB2019-055. Epub 2019 May 10. Cancer Discov. 2019. PMID: 31076483
Similar articles
-
Discovery of WRN inhibitor HRO761 with synthetic lethality in MSI cancers.Nature. 2024 May;629(8011):443-449. doi: 10.1038/s41586-024-07350-y. Epub 2024 Apr 24. Nature. 2024. PMID: 38658754 Free PMC article.
-
WRN Is a Promising Synthetic Lethal Target for Cancers with Microsatellite Instability (MSI).Cancer Treat Res. 2023;186:313-328. doi: 10.1007/978-3-031-30065-3_17. Cancer Treat Res. 2023. PMID: 37978143
-
Repeat expansions confer WRN dependence in microsatellite-unstable cancers.Nature. 2020 Oct;586(7828):292-298. doi: 10.1038/s41586-020-2769-8. Epub 2020 Sep 30. Nature. 2020. PMID: 32999459 Free PMC article.
-
The WRN helicase: resolving a new target in microsatellite unstable cancers.Curr Opin Genet Dev. 2021 Dec;71:34-38. doi: 10.1016/j.gde.2021.06.014. Epub 2021 Jul 17. Curr Opin Genet Dev. 2021. PMID: 34284257 Review.
-
Roles of Caenorhabditis elegans WRN Helicase in DNA Damage Responses, and a Comparison with Its Mammalian Homolog: A Mini-Review.Gerontology. 2016;62(3):296-303. doi: 10.1159/000439200. Epub 2015 Sep 9. Gerontology. 2016. PMID: 26347143 Review.
Cited by
-
Clinical prospects of WRN inhibition as a treatment for MSI tumours.NPJ Precis Oncol. 2022 Nov 15;6(1):85. doi: 10.1038/s41698-022-00319-y. NPJ Precis Oncol. 2022. PMID: 36379964 Free PMC article. Review.
-
The USP1 Inhibitor KSQ-4279 Overcomes PARP Inhibitor Resistance in Homologous Recombination-Deficient Tumors.Cancer Res. 2024 Oct 15;84(20):3419-3434. doi: 10.1158/0008-5472.CAN-24-0293. Cancer Res. 2024. PMID: 39402989 Free PMC article.
-
VRK1 Is a Synthetic-Lethal Target in VRK2-Deficient Glioblastoma.Cancer Res. 2022 Nov 2;82(21):4044-4057. doi: 10.1158/0008-5472.CAN-21-4443. Cancer Res. 2022. PMID: 36069976 Free PMC article.
-
Spectrum of DNA mismatch repair failures viewed through the lens of cancer genomics and implications for therapy.Clin Sci (Lond). 2022 Mar 18;136(5):383-404. doi: 10.1042/CS20210682. Clin Sci (Lond). 2022. PMID: 35274136 Free PMC article.
-
Inactivating Mutations in Exonuclease and Polymerase Domains in DNA Polymerase Delta Alter Sensitivities to Inhibitors of dNTP Synthesis.DNA Cell Biol. 2020 Jan;39(1):50-56. doi: 10.1089/dna.2019.5125. Epub 2019 Nov 21. DNA Cell Biol. 2020. PMID: 31750734 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

