New tools for automated high-resolution cryo-EM structure determination in RELION-3
- PMID: 30412051
- PMCID: PMC6250425
- DOI: 10.7554/eLife.42166
New tools for automated high-resolution cryo-EM structure determination in RELION-3
Abstract
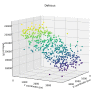
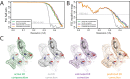
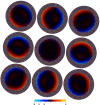
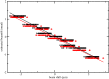
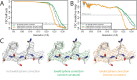
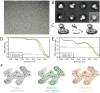
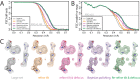
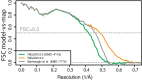
Here, we describe the third major release of RELION. CPU-based vector acceleration has been added in addition to GPU support, which provides flexibility in use of resources and avoids memory limitations. Reference-free autopicking with Laplacian-of-Gaussian filtering and execution of jobs from python allows non-interactive processing during acquisition, including 2D-classification, de novo model generation and 3D-classification. Per-particle refinement of CTF parameters and correction of estimated beam tilt provides higher resolution reconstructions when particles are at different heights in the ice, and/or coma-free alignment has not been optimal. Ewald sphere curvature correction improves resolution for large particles. We illustrate these developments with publicly available data sets: together with a Bayesian approach to beam-induced motion correction it leads to resolution improvements of 0.2-0.7 Å compared to previous RELION versions.
Keywords: Cryo-EM; molecular biophysics; none; single-particle analysis; software; structural biology.
© 2018, Zivanov et al.
Conflict of interest statement
JZ, TN, BF, DK, WH, EL No competing interests declared, SS Reviewing editor, eLife
Figures
Similar articles
-
A Robust Single-Particle Cryo-Electron Microscopy (cryo-EM) Processing Workflow with cryoSPARC, RELION, and Scipion.J Vis Exp. 2022 Jan 31;(179). doi: 10.3791/63387. J Vis Exp. 2022. PMID: 35104261
-
Accelerated cryo-EM structure determination with parallelisation using GPUs in RELION-2.Elife. 2016 Nov 15;5:e18722. doi: 10.7554/eLife.18722. Elife. 2016. PMID: 27845625 Free PMC article.
-
A Bayesian approach to single-particle electron cryo-tomography in RELION-4.0.Elife. 2022 Dec 5;11:e83724. doi: 10.7554/eLife.83724. Elife. 2022. PMID: 36468689 Free PMC article.
-
Processing of Structurally Heterogeneous Cryo-EM Data in RELION.Methods Enzymol. 2016;579:125-57. doi: 10.1016/bs.mie.2016.04.012. Epub 2016 May 31. Methods Enzymol. 2016. PMID: 27572726 Review.
-
High resolution single particle Cryo-EM refinement using JSPR.Prog Biophys Mol Biol. 2021 Mar;160:37-42. doi: 10.1016/j.pbiomolbio.2020.05.006. Epub 2020 Jul 3. Prog Biophys Mol Biol. 2021. PMID: 32622834 Free PMC article. Review.
Cited by
-
Helical ultrastructure of the metalloprotease meprin α in complex with a small molecule inhibitor.Nat Commun. 2022 Oct 19;13(1):6178. doi: 10.1038/s41467-022-33893-7. Nat Commun. 2022. PMID: 36261433 Free PMC article.
-
Ctf3/CENP-I provides a docking site for the desumoylase Ulp2 at the kinetochore.J Cell Biol. 2021 Aug 2;220(8):e202012149. doi: 10.1083/jcb.202012149. Epub 2021 Jun 3. J Cell Biol. 2021. PMID: 34081091 Free PMC article.
-
Molecular mechanism underlying transport and allosteric inhibition of bicarbonate transporter SbtA.Proc Natl Acad Sci U S A. 2021 Jun 1;118(22):e2101632118. doi: 10.1073/pnas.2101632118. Proc Natl Acad Sci U S A. 2021. PMID: 34031249 Free PMC article.
-
Structure of the Inhibited State of the Sec Translocon.Mol Cell. 2020 Aug 6;79(3):406-415.e7. doi: 10.1016/j.molcel.2020.06.013. Epub 2020 Jul 9. Mol Cell. 2020. PMID: 32692975 Free PMC article.
-
Assessing the JEOL CRYO ARM 300 for high-throughput automated single-particle cryo-EM in a multiuser environment.IUCrJ. 2020 Jun 11;7(Pt 4):707-718. doi: 10.1107/S2052252520006065. eCollection 2020 Jul 1. IUCrJ. 2020. PMID: 32695417 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

