Targeting endothelin receptor signalling overcomes heterogeneity driven therapy failure
- PMID: 28606996
- PMCID: PMC5538298
- DOI: 10.15252/emmm.201607156
Targeting endothelin receptor signalling overcomes heterogeneity driven therapy failure
Abstract
Approaches to prolong responses to BRAF targeting drugs in melanoma patients are challenged by phenotype heterogeneity. Melanomas of a "MITF-high" phenotype usually respond well to BRAF inhibitor therapy, but these melanomas also contain subpopulations of the de novo resistance "AXL-high" phenotype. > 50% of melanomas progress with enriched "AXL-high" populations, and because AXL is linked to de-differentiation and invasiveness avoiding an "AXL-high relapse" is desirable. We discovered that phenotype heterogeneity is supported during the response phase of BRAF inhibitor therapy due to MITF-induced expression of endothelin 1 (EDN1). EDN1 expression is enhanced in tumours of patients on treatment and confers drug resistance through ERK re-activation in a paracrine manner. Most importantly, EDN1 not only supports MITF-high populations through the endothelin receptor B (EDNRB), but also AXL-high populations through EDNRA, making it a master regulator of phenotype heterogeneity. Endothelin receptor antagonists suppress AXL-high-expressing cells and sensitize to BRAF inhibition, suggesting that targeting EDN1 signalling could improve BRAF inhibitor responses without selecting for AXL-high cells.
Keywords: AXL; BRAF; MITF; endothelin; melanoma.
© 2017 The Authors. Published under the terms of the CC BY 4.0 license.
Figures
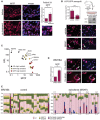
Immunofluorescence analysis for MITF (magenta) in a tumour of a patient, who had been treated with dabrafenib for 2 weeks. Nuclei were stained with DAPI. Scale bar: 10 μm. Relative MITF mRNA expression assessed by qRT–PCR is shown. P: probability by paired t‐test: *P = 0.0219. Error bars: SD of three replicate measures.
A375‐GFP cells were isolated from xenografts grown in mice (n = 6) that had been treated with vehicle or 100 mg/kg vemurafenib (BRAFi) for 12 days (maximum response). The tumour volume is indicated (mean ± SD; horizontal black line, 100 mm3, volume at start); P: probability by t‐test: ***P = 0.001. The ex vivo cultures were analysed for MITF expression by Western blot and immunofluorescence (magenta). Nuclei were stained with DAPI. Scale bar: 20 μm (white arrows, high MITF; black arrows, low MITF).
Relative AXL and MITF expression in a panel of melanoma cell lines that have been characterized for their response to BRAF inhibition (Barretina et al, 2012; Garnett et al, 2012; Smith et al, 2013).
MITF immunofluorescence analysis of WM164 cells treated with DMSO or dabrafenib for 72 h. Scale bar: 10 μm. Relative MITF mRNA expression assessed by qRT–PCR is shown (n = 3 independent experiments; mean ± SEM). P: probability by paired t‐test: **P = 0.0095 (white arrows, high MITF; black arrows, low MITF).
Time‐lapse analysis of mKO2‐hCdt1 and mAG‐hGem (FUCCI) expressing WM164 melanoma cells (Haass et al, 2014) over 72 h. Cells were either treated with DMSO or dabrafenib. Dashed lines indicate cells whose daughter cells underwent different fates after exiting mitosis.
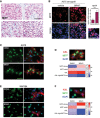
Immunohistochemistry analysis of MITF expression in human melanoma specimens. Scale bar: 100 μm.
Immunofluorescence analysis of MITF expression (magenta) in xenografts from mice treated with either vehicle or 25 mg/kg PLX4720 (BRAFi) for 3 weeks. Nuclei were stained with DAPI. Scale bar: 100 μm. Dashed line indicates origin of magnification; scale bar: 50 μm. RT‐qPCR for MITF from xenografts (n = 5) is shown. *** indicates probability by t‐test; P < 0.0001. Data are presented as mean ± SEM.
Immunofluorescence analysis in A375 melanoma cells treated with either DMSO or vemurafenib for 3 days, before they were stained for MITF (green) and AXL (red) expression. Scale bar: 50 μm.
Quantification of MITF and AXL co‐staining in single cells. Scale bar: 50 μm.
Immunofluorescence analysis in WM164 melanoma cells treated with either DMSO or vemurafenib for 3 days, before they were stained for MITF (green) and AXL (red) expression. Scale bar: 50 μm.
Quantification of MITF and AXL co‐staining in single cells. Scale bar: 50 μm.
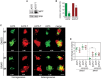
Western blot of A375 or A375‐T cells depicting basal expression of MITF. ERK2 served as loading control.
A375 or A375‐T cells were treated with vemurafenib (BRAFi) for 72 h before relative cell number was assessed. A Western blot for MITF in A375 and A375‐T cells is shown. P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001 (A375) and **P = 0.0022 (A375‐T).
GFP‐expressing A375 or RFP‐expressing A375‐T cells were injected into the pericardial space of zebrafish embryos, before they were treated with either vemurafenib (BRAFi) or DMSO. The total number of cells for each injection condition was 1,000 cells. Xenografts were imaged at day 1 and day 4 of drug treatment using a Leica SP5 confocal microscope, and fold change in volume of populations of GFP‐ or RFP‐expressing cells at day 4 compared to day 1 is indicated. Scale bar: 100 μm.
Xenograft volumes seen in 3D images at day 1 and day 4 of treatment were quantified using Volocity® software. Fold change relative to day 1 is shown. P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001.
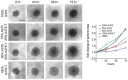

Dose–response curves for vemurafenib (BRAFi) in the indicated cell lines. The respective cell lines were treated with either DMEM (control) or conditioned medium, which was derived from the respective cell line treated with vemurafenib for the indicated times. Western blots for MITF with ERK2 as loading control are shown.
Schematic of co‐culture assay of drug‐tolerant and drug‐sensitive melanoma cells. Drug‐sensitive cells were co‐cultured with either drug‐sensitive cells or cells that had been pre‐treated with vemurafenib for 14 days. The co‐cultures were treated for 48 h with vemurafenib (BRAFi) before quantitative analysis of paracrine protection. Paracrine protection was determined as difference between co‐culture with sensitive cells and drug pre‐treated cells. P: probability by one‐way ANOVA (with Tukey's post hoc test); **P = 0.0022 (A375), *P = 0.0307 (M249), **P = 0.0084 (WM9) and ***P < 0.0001 (WM164).
Analysis of paracrine protection in a panel of drug‐tolerant and drug‐sensitive melanoma cells. Drug‐sensitive A375, WM9 or WM98 cells were co‐cultured with the indicated melanoma cell cultures, and paracrine protection was determined as indicated in (B).
Western blot of the indicated cell cultures for pERK and total ERK. The indicated cultures were co‐cultured with either untreated cells (control) or with the respective drug pre‐treated cell lines. Cells were treated with DMSO or with vemurafenib (BRAFi) for 24 h.
Western blot of the indicated cell cultures for pERK and total ERK. Cells were either treated with DMSO or treated with vemurafenib (BRAFi) in the presence or absence of RAF265 or GO‐6983 (PKCi).
Quantification of relative cell numbers. A375 cells were co‐cultured with either A375 or with A375‐T cells. DMSO‐treated A375 cells in the presence of A375 cells were set at 100%. P: probability by t‐test: ns P > 0.05, ***P < 0.0001 and *P = 0.0312.
Western blot for pERK and proteins that represent PKC substrates. ERK served as loading control. The indicated cell lines had been either left untreated or were treated with conditioned medium derived from the respective drug pre‐treated (14 days) cell lines.
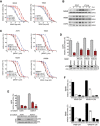
Dose–response curves for vemurafenib (BRAFi) for the indicated cell lines. The cell lines were treated in either DMEM (control) or conditioned medium, which was derived from cells treated with vemurafenib for the indicated times.
Western blot for MITF in the indicated cell lines treated with vemurafenib (BRAFi) for the indicated times.
Dose–response curves for vemurafenib (BRAFi) for the indicated cell lines. The cell lines were treated in either DMEM (control) or conditioned medium, which was derived from cells treated with vemurafenib, and then, the inhibitor was removed for the indicated times.
Quantification of relative cell numbers. A375 cells were either left untreated or were treated with 0.5 μM vemurafenib (BRAFi) in the presence of conditioned medium from ex vivo cultures. DMSO‐treated A375 cells were set at 100%. A Western blot for pERK and ERK under the respective conditions is shown. P: probability by one‐way ANOVA (with Dunnett's post hoc test); **P = 0.0014 (#867), **P = 0.0026 (#549) and *P = 0.0387 (#026).
Western blots of A375 cells for pERK and total ERK, and quantification of relative cell number of A375 cells when co‐cultured with either A375 or with A375‐T cells. Cells were treated with DMSO or vemurafenib (BRAFi) or selumetinib (MEKi) for 48 h. A375 cells in the presence of A375 cells were set at 100%. P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, **P = 0.001.
Quantification of relative cell number of the indicated cell lines treated with DMSO or with vemurafenib (BRAFi). During drug treatment, cells were incubated with conditioned medium derived from untreated cells or cells pre‐treated with BRAFi for 14 days. P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001.
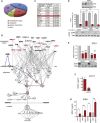
Schematic presentation of groups of proteins detected by quantitative mass spectrometry to be enriched in conditioned medium of A375‐T cells when compared to A375 cells.
Ingenuity Pathway Analysis of factors in the extracellular space for involvement in the activation of ERK. MITF targets are indicated in red font.
Ingenuity Upstream Regulator Analysis shows the potential transcriptional regulators that can explain changes observed in secreted proteins. Proteins that were up‐regulated in the media of drug‐treated cells (FC > 2, FDR < 0.05) were selected for IPA analysis, and upstream transcriptional driver were identified. The Z‐score indicates activation states of predicted regulators with positive values corresponding to activated transcription regulator activating gene expression.
Quantification of relative cell number and analysis of ERK phosphorylation of A375 cells when co‐cultured with either A375 or with A375‐T cells. DMSO‐treated A375 cells in the presence of A375 cells = 100%. Before co‐culture, A375‐T cells were treated with a control or two different MITF‐specific siRNAs (siMI1, siMI3). A Western blot demonstrating the degree of MITF knockdown is shown. ERK served as loading control. P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, **P = 0.0047 (A375 co‐culture vs. A375‐T co‐culture).
qRT–PCR for ECE1 expression in the indicated melanoma cell lines treated with a control (sicon) or two different MITF‐specific siRNAs (siMI1, siMI3). A Western blot demonstrating the degree of MITF knockdown is shown. ERK served as loading control. P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001 (A375‐T siMi1 and siMi3, A375‐T2 siMi1), ***P = 0.0003 (A375T2‐siMI3).
A375‐T cells were left untreated or treated for 24 h with 15 μM ECE1 inhibitor CGS 35066 before EDN1 levels in the medium were analysed by ELISA. P: probability by t‐test: ***P < 0.0001.
ELISA measuring EDN1 concentrations in the medium of the indicated cell lines. Cells were either untreated or had been treated with BRAFi for 14 days. P: probability by one‐way ANOVA (with Sidak's post hoc test); ***P < 0.0001 (WM9, M249), **P = 0.0013 (WM98) and *P = 0.0238 (A375).
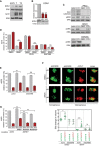
Western blot for EDN1 and MITF expression in A375, A375‐T and A375‐T2 cells. Beta‐tubulin and ERK2 served as loading control, respectively.
qRT–PCR for EDN1 expression and Western blot for EDN1 in A375‐T cells after treatment with either a control siRNA (sicon) or two different MITF‐specific siRNAs (siMI1, SiMI3). P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001.
A375 cells were stimulated with EDN1 for 8 h and analysed for phosphorylation of PKC substrates, pERK and total ERK on a Western blot. Cells were treated with DMSO or GO‐6983 (PKCi) before analysis (upper panel), or vemurafenib (BRAFi, middle panel) or RAF265 (lower panel).
Quantification of relative cell number of A375 cells when treated with EDN1 in the absence or presence of vemurafenib (BRAFi) and GO‐6983 (PKCi) or RAF265. P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, ***P = 0.0006.
Quantification of relative cell number of A375‐pLKO or A375‐shEDNRB cells when treated with EDN1 in the absence or presence of BRAFi. P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, ***P = 0.0003.
GFP‐A375 pLKO, GFP‐A375 shEDNRB or RFP‐A375‐T cells were injected into the pericardial space of zebrafish embryos, and the embryos were treated with either vemurafenib (BRAFi) or the vehicle DMSO. Images at day 1 and day 4 of treatment are shown, and fold change in volume at day 4 compared to day 1 was quantified. Scale bar: 100 μm. P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, *P = 0.026 (DMSO), ***P < 0.0001 (BRAFi).
Quantification of relative cell number of A375 cells when co‐cultured with either A375 or with A375‐T cells in the absence or presence of BRAFi, alone or in combination with bosentan or macitentan. P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001 (all).

Quantification of relative cell number of A375 cells when treated with EDN1 in the absence or presence of vemurafenib (BRAFi) and selumetinib (MEKi).
Quantification of relative cell number of A375 cells when co‐cultured with A375‐T cells treated with a control or an EDN1 specific siRNA. P: probability by one‐way ANOVA (with Tukey's post hoc test); **P = 0.0011. A Western blot demonstrating the degree of EDN1 knock down in A375‐T cells is shown. Beta‐tubulin served as loading control.
Quantification of relative cell number of A375 cells when co‐cultured with A375‐T cells in the presence of a control or an EDN1‐specific blocking antibody. P: probability by one‐way ANOVA (with Tukey's post hoc test); **P = 0.0016.
The indicated cancer cell line datasets were analysed for EDNRB expression using Oncomine. P: probability by t‐test; ***P < 0.0001 (Garnett and Barretina). Mean ± SD; n = 732 (Garnett); n = 917 (Barretina).
BRAFi dose–response curves for vemurafenib (BRAFi) or the indicated cell lines. The cell lines were treated in DMEM (control) or in conditioned medium derived from cells treated for 14 days with BRAFi in the absence or presence of bosentan.
Real‐time qPCR of EDN1 in short‐term cultures from progressed patients.
Analysis of paracrine protection in a panel of short‐term cultures from progressed patients and drug‐sensitive melanoma cells. Drug‐sensitive A375, WM9 or WM98 cells were co‐cultured with the indicated melanoma cell cultures, and paracrine detection was determined as indicated. Data are the mean of three independent experiments.
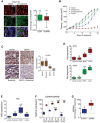
- A
Analysis of EDN1 and EDNRB expression in patients. Immunofluorescence analysis for MITF and EDN1 expression in the tumour of patient 24 before and on treatment. Scale bar: 10 μm. EDN1 and EDNRB qRT–PCR analysis in tumours of patients on treatment with either vemurafenib alone or a dabrafenib/trametinib combination (n = 22). P: probability by t‐test; **P = 0.0064 (EDN1) and **P = 0.0033 (EDNRB).
- B
Nude mice bearing A375 tumours were treated (n = 5–6 mice per group) with vehicle, vemurafenib (25 mg/kg/qd) or bosentan (25 mg/kg/qd) alone or in combination for 20 days. Data are presented as mean tumour volumes ± SEM.
- C
Phospho‐ERK IHC and qRT–PCR for DUSP6 from tumours corresponding to the experiment described in (B). Scale bar: 200 μm. P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, *P = 0.0186 (BRAFi) and ***P = 0.0004 (combo).
- D
qRT–PCR for MITF and EDN1 from tumours corresponding to the experiment described in (B). P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, *P = 0.0179 (MITF‐BRAFi), ***P < 0.0001 (MITF‐combo), ***P < 0.0001 (EDN1‐BRAFi) and ***P < 0.0001 (EDN1‐combo).
- E, F
qRT–PCR for AXL, and EDNRA and EDNRB from tumours corresponding to the experiment described in (B). P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, *P = 0.0341 (EDNRA‐Bosentan) and ***P < 0.0001 (AXL‐BRAFi, EDNRA‐BRAFi, EDNRB‐BRAFi and EDNRB‐combo).
- G
qRT–PCR for EDNRA and EDNRB in tumours of patients on treatment with either vemurafenib alone or a dabrafenib/trametinib combination. Relative basal expression of EDNRA and EDNRB was considered. P: probability by t‐test; *P = 0.0116 (EDNRA) and **P = 0.0039 (EDNRB).

Correlation analysis for the expression of the indicated genes using the TCGA‐melanoma dataset (TCGANetwork, 2015).
Real‐time qPCR of EDNRA and EDNRB in the indicated MITF‐high and AXL‐high cell lines.
Analysis of cells in S‐phase. The indicated cell lines were treated with vemurafenib and 20 ng/ml EDN1 either alone or in combination for 24 h, and 4 h before analysis EdU was added to the cultures. P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001 (WM266‐4, WM164, WM793), **P = 0.009 (A375), *P = 0.0445 (RPMI7951).
Analysis of cells in S‐phase. The indicated cell lines were treated with vemurafenib (BRAFi), BQ788 or BQ123 either alone or in combination and in the presence of conditioned medium from A375‐T cells for 24 h, and 4 h before analysis EdU was added to the cultures. The conditioned medium from A375 cells was used as control. P: probability by t‐test; ns P > 0.05, ***P = 0.0007 (A375, BRAFi/BQ788) and ***P = 0.0009 (WM793, BRAFi/BQ123).
Western blot of A375 and WM793 cells in the absence or presence of conditioned medium from A375‐T cells treated with DMSO or BRAF inhibitor for phospho‐ERK and PKC substrates as indicated. ERK2 served as loading control.
Quantification of relative cell numbers. A375 or WM793 cells were either left untreated or were treated with 0.5 μM vemurafenib (BRAFi) and bosentan either alone or in combination in the presence of conditioned medium from A375‐T cells for one week. DMSO‐treated A375 cells were set at 100%. P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001.
Western blot for the indicated proteins of the indicated cell lines with DMSO, vemurafenib (BRAFi) or BRAFi in the presence of conditioned medium from A375‐T cells.
Analysis for apoptosis using an incucyte® caspase‐3/7 apoptosis assay reagent. The indicated cell lines were treated with DMSO, vemurafenib (BRAFi) alone or with BRAFi +/− macitentan (Mac) in the presence of conditioned medium from A375‐T cells and apoptosis activity was measured over time; end‐point values at 48 h are shown. P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, ***P < 0.0001.

Pearson correlation of expression (log2) of EDNRA with AXL in melanoma cell lines from the Barretina and Garnett (Garnett et al, 2012) datasets deposited in Oncomine.
Pearson correlation of expression (log2) of EDNRB with either the expression (log2) of EDNRA or AXL in melanoma cell lines from the Barretina dataset (Barretina et al, 2012) deposited in Oncomine.
Analysis of cells in S‐phase. The indicated cell lines were treated with 0.5 μM vemurafenib (BRAFi) and bosentan either alone or in combination and in the presence of conditioned medium from A375‐T cells for 24 h, and 4 h before analysis EdU was added to the cultures. P: probability by t‐test; ***P < 0.0001 (A375), ***P < 0.0001 (WM164), ***P < 0.0001 (WM793) and *P = 0.0445 (RPMI7951).
Western blot for pERK and ERK of A375 and WM793 cells treated with vemurafenib (BRAFi) and bosentan either alone or in combination and in the presence of conditioned medium.
Quantification of relative cell number of A375 cells when co‐cultured with either A375 or with A375‐T cells in the absence or presence of vemurafenib (BRAFi), alone or in combination with BQ788 or BQ123. P: probability by one‐way ANOVA (with Dunnett's post hoc test); ns P > 0.05, ***P < 0.0001.
Quantification of relative cell number of A375 and WM793 cells treated with vemurafenib (BRAFi) and macitentan or BQ788 or BQ123 either alone or in combination and in the presence of conditioned medium. P: probability by one‐way ANOVA (with Dunnett's post hoc test); ns P > 0.05, ***P < 0.0001 (WM793 BQ123 + BRAFi, A375 Mac + BRAFi, BQ788 + BRAFi), ***P = 0.0004 (WM793 Mac + BRAFi).
Quantification of relative cell number of WM793 cells when co‐cultured with A375‐T cells in the presence of a control or an EDN1‐specific blocking antibody. Conditioned medium from A375 cells was used as control. ns P > 0.05, ***P = 0.0005; Tukey's test.
Quantification of relative cell number of WM793 cells treated with EDN1 in the absence or presence of vemurafenib (BRAFi), alone or in combination with RAF265 or dovitinib (RTKi) or GO‐6983 (PKCi). P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, ***P < 0.0001 (RAF265 + BRAFi, PKCi + BRAFi).
Western blot for pERK and ERK2 of WM793 cells when treated with EDN1 in the presence of vemurafenib (BRAFi) and RAF265, dovitinib (RTKi) or GO‐6983 (PKCi).
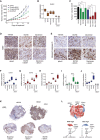
- A
Nude mice bearing A375 tumours were treated (n = 5–6 mice per group) with vehicle, vemurafenib (25 mg/kg/qd) or BQ788 (10 mg/kg/qd) alone or in combination for 20 days.
- B
qRT–PCR for DUSP6 from tumours corresponding to the experiment described in (A) and Fig EV6A. P: probability by one‐way ANOVA (with Tukey's post hoc test); ***P < 0.0001 (BRAFi, BRAFi + Macitentan, BRAFi + BQ788), **P = 0.068 (Macitentan) and **P = 0.082 (BQ788). Error bars, min and max values; box limits, second and third quartiles; horizontal line, median.
- C
A375 tumour volume on day 18 of treatment with the indicated regimes. P: probability by one‐way ANOVA (with Tukey's post hoc test); ns P > 0.05, ***P = 0.0004 (BQ788), ***P < 0.0001 (Bos + BRAFi, BQ + BRAFi), **P = 0.009 (Mac + BRAFi).
- D
IHC for Ki67 in A375 xenografts from mice treated as indicated. Scale bar: 50 μm.
- E
IHC for cleaved caspase‐3 in A375 xenografts from mice treated as indicated. Scale bar: 50 μm.
- F–J
qRT–PCR for EDNRA, EDNRB, MITF, EDN1 and AXL expression in tumours corresponding to the experiment described in (A) and Fig EV6A. P: probability by one‐way ANOVA (with Tukey's post hoc test); (F) EDNRA: ns P > 0.05, **P = 0.0010 (BQ788), ***P < 0.0001 (BRAFi) and ***P < 0.0001 (BQ788 + BRAFi). (G) EDNRB: ns P > 0.05, ***P < 0.0001 (BRAFi), ***P = 0.0002 (Mac + BRAFi) and ***P < 0.0001 (BQ788 + BRAFi). (H) MITF: ns P > 0.05, ***P < 0.0001 (BRAFi), ***P = 0.0001 (Mac + BRAFi), and ***P < 0.0001 (BQ788 + BRAFi). (I) EDN1: ns P > 0.05, ***P < 0.0001 (BRAFi), **P = 0.0331 (Mac + BRAFi) and **P = 0.0225 (BQ788 + BRAFi). (J) AXL: ns P > 0.05, *P = 0.0462 (BQ788) and ***P < 0.0001 (BRAFi).
- K
IHC for AXL of A375 xenografts from mice treated as indicated. Scale bar: 2,000 μm.
- L
Model of EDN1‐mediated paracrine protection. EDN1 induces re‐activation ERK in the presence of BRAF inhibitor. In MITF‐high cells, ERK regulates proliferation and survival, but in AXL‐high cells ERK only stimulate proliferation.
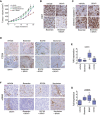
Nude mice (n = 5–6 mice per group) bearing A375 tumours were treated with vehicle, vemurafenib (25 mg/kg/qd) or macitentan (10 mg/kg/qd) alone or in combination for 20 days.
IHC for Ki67 in A375 xenografts from mice treated as indicated. Scale bar: 50 μm.
IHC for cleaved caspase‐3 in A375 xenografts from mice treated as indicated. Scale bar: 50 μm.
IHC for cleaved CD34 in A375 xenografts from mice treated as indicated. Scale bar: 100 μm.
qRT–PCR for CD31 from tumours treated with BRAF inhibitor or bosentan alone or in combination. P: probability by one‐way ANOVA (with Dunnett's post hoc test); ns P > 0.05, **P = 0.0012. Error bars, min and max values; box limits, second and third quartiles; horizontal line, median.
IHC for cleaved αSMA in A375 xenografts from mice treated as indicated. Scale bar: 50 μm.
qRT–PCR for αSMA from tumours treated with BRAF inhibitor or bosentan alone or in combination. P: probability by one‐way ANOVA (with Dunnett's post hoc test); ns P > 0.05, *P = 0.0255 (Bosentan) and **P = 0.0060 (BRAFi). Error bars, min and max values; box limits, second and third quartiles; horizontal line, median.
Comment in
-
Phenotypic diversity drives paracrine drug tolerance.EMBO Mol Med. 2017 Aug;9(8):987-989. doi: 10.15252/emmm.201707956. EMBO Mol Med. 2017. PMID: 28694322 Free PMC article.
Similar articles
-
Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma.Nat Commun. 2014 Dec 15;5:5712. doi: 10.1038/ncomms6712. Nat Commun. 2014. PMID: 25502142 Free PMC article.
-
A melanoma cell state distinction influences sensitivity to MAPK pathway inhibitors.Cancer Discov. 2014 Jul;4(7):816-27. doi: 10.1158/2159-8290.CD-13-0424. Epub 2014 Apr 25. Cancer Discov. 2014. PMID: 24771846 Free PMC article.
-
AXL/AKT axis mediated-resistance to BRAF inhibitor depends on PTEN status in melanoma.Oncogene. 2018 Jun;37(24):3275-3289. doi: 10.1038/s41388-018-0205-4. Epub 2018 Mar 19. Oncogene. 2018. PMID: 29551771 Free PMC article.
-
Vemurafenib in patients with BRAF V600E mutation-positive advanced melanoma.Clin Ther. 2012 Jul;34(7):1474-86. doi: 10.1016/j.clinthera.2012.06.009. Epub 2012 Jun 27. Clin Ther. 2012. PMID: 22742884 Review.
-
Targeting BRAF in melanoma: biological and clinical challenges.Crit Rev Oncol Hematol. 2013 Sep;87(3):239-55. doi: 10.1016/j.critrevonc.2013.01.003. Epub 2013 Feb 15. Crit Rev Oncol Hematol. 2013. PMID: 23415641 Review.
Cited by
-
The EDN1/EDNRA/β‑arrestin axis promotes colorectal cancer progression by regulating STAT3 phosphorylation.Int J Oncol. 2023 Jan;62(1):13. doi: 10.3892/ijo.2022.5461. Epub 2022 Dec 1. Int J Oncol. 2023. PMID: 36453252 Free PMC article.
-
Many Distinct Ways Lead to Drug Resistance in BRAF- and NRAS-Mutated Melanomas.Life (Basel). 2021 May 5;11(5):424. doi: 10.3390/life11050424. Life (Basel). 2021. PMID: 34063141 Free PMC article. Review.
-
Melanoma: Genetic Abnormalities, Tumor Progression, Clonal Evolution and Tumor Initiating Cells.Med Sci (Basel). 2017 Nov 20;5(4):28. doi: 10.3390/medsci5040028. Med Sci (Basel). 2017. PMID: 29156643 Free PMC article. Review.
-
HDAC Inhibition Enhances the In Vivo Efficacy of MEK Inhibitor Therapy in Uveal Melanoma.Clin Cancer Res. 2019 Sep 15;25(18):5686-5701. doi: 10.1158/1078-0432.CCR-18-3382. Epub 2019 Jun 21. Clin Cancer Res. 2019. PMID: 31227503 Free PMC article.
-
Stromal Cells Present in the Melanoma Niche Affect Tumor Invasiveness and Its Resistance to Therapy.Int J Mol Sci. 2021 Jan 7;22(2):529. doi: 10.3390/ijms22020529. Int J Mol Sci. 2021. PMID: 33430277 Free PMC article. Review.
References
-
- Beaumont KA, Hill DS, Daignault SM, Lui GY, Sharp DM, Gabrielli B, Weninger W, Haass NK (2016) Cell cycle phase‐specific drug resistance as an escape mechanism of melanoma cells. J Invest Dermatol 136: 1479–1489 - PubMed
-
- Boss C, Bolli MH, Gatfield J (2016) From bosentan (Tracleer(R)) to macitentan (Opsumit(R)): the medicinal chemistry perspective. Bioorg Med Chem Lett 26: 3381–3394 - PubMed
-
- Carlino MS, Long GV, Kefford RF, Rizos H (2015) Targeting oncogenic BRAF and aberrant MAPK activation in the treatment of cutaneous melanoma. Crit Rev Oncol Hematol 96: 385–398 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

