Assembly of functionally integrated human forebrain spheroids
- PMID: 28445465
- PMCID: PMC5805137
- DOI: 10.1038/nature22330
Assembly of functionally integrated human forebrain spheroids
Abstract
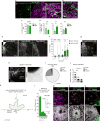
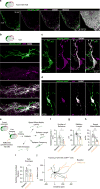
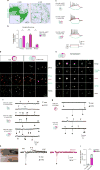
The development of the nervous system involves a coordinated succession of events including the migration of GABAergic (γ-aminobutyric-acid-releasing) neurons from ventral to dorsal forebrain and their integration into cortical circuits. However, these interregional interactions have not yet been modelled with human cells. Here we generate three-dimensional spheroids from human pluripotent stem cells that resemble either the dorsal or ventral forebrain and contain cortical glutamatergic or GABAergic neurons. These subdomain-specific forebrain spheroids can be assembled in vitro to recapitulate the saltatory migration of interneurons observed in the fetal forebrain. Using this system, we find that in Timothy syndrome-a neurodevelopmental disorder that is caused by mutations in the CaV1.2 calcium channel-interneurons display abnormal migratory saltations. We also show that after migration, interneurons functionally integrate with glutamatergic neurons to form a microphysiological system. We anticipate that this approach will be useful for studying neural development and disease, and for deriving spheroids that resemble other brain regions to assemble circuits in vitro.
Conflict of interest statement
H. C.F. and K.C.M. were employees of BD Genomics during this study.
Figures



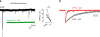






Comment in
-
Human development: Advances in mini-brain technology.Nature. 2017 May 3;545(7652):39-40. doi: 10.1038/545039a. Nature. 2017. PMID: 28470206 No abstract available.
-
Modeling human brain development.Sci Transl Med. 2017 Jun 7;9(393):eaan4295. doi: 10.1126/scitranslmed.aan4295. Sci Transl Med. 2017. PMID: 28592564
-
You Have Brains in Your Head, You Have Organoids in Your Dish, You Can Steer Yourself in any Direction You Wish.Epilepsy Curr. 2017 Sep-Oct;17(5):311-313. doi: 10.5698/1535-7597.17.5.311. Epilepsy Curr. 2017. PMID: 29225549 Free PMC article. No abstract available.
Similar articles
-
Generation and assembly of human brain region-specific three-dimensional cultures.Nat Protoc. 2018 Sep;13(9):2062-2085. doi: 10.1038/s41596-018-0032-7. Nat Protoc. 2018. PMID: 30202107 Free PMC article.
-
Direct Induction and Functional Maturation of Forebrain GABAergic Neurons from Human Pluripotent Stem Cells.Cell Rep. 2016 Aug 16;16(7):1942-53. doi: 10.1016/j.celrep.2016.07.035. Epub 2016 Aug 4. Cell Rep. 2016. PMID: 27498872
-
Dissecting the molecular basis of human interneuron migration in forebrain assembloids from Timothy syndrome.Cell Stem Cell. 2022 Feb 3;29(2):248-264.e7. doi: 10.1016/j.stem.2021.11.011. Epub 2022 Jan 5. Cell Stem Cell. 2022. PMID: 34990580
-
Distinct origin of GABA-ergic neurons in forebrain of man, nonhuman primates and lower mammals.Coll Antropol. 2008 Jan;32 Suppl 1:9-17. Coll Antropol. 2008. PMID: 18405052 Review.
-
Unusual clinical description of adult with Timothy syndrome, carrier of a new heterozygote mutation of CACNA1C.Eur J Med Genet. 2019 Jul;62(7):103648. doi: 10.1016/j.ejmg.2019.04.005. Epub 2019 Apr 16. Eur J Med Genet. 2019. PMID: 30998997 Review.
Cited by
-
Single-cell sequencing technology in skin wound healing.Burns Trauma. 2024 Oct 23;12:tkae043. doi: 10.1093/burnst/tkae043. eCollection 2024. Burns Trauma. 2024. PMID: 39445224 Free PMC article. Review.
-
Organoid intelligence for developmental neurotoxicity testing.Front Cell Neurosci. 2024 Oct 8;18:1480845. doi: 10.3389/fncel.2024.1480845. eCollection 2024. Front Cell Neurosci. 2024. PMID: 39440004 Free PMC article.
-
Bioprinting of Cells, Organoids and Organs-on-a-Chip Together with Hydrogels Improves Structural and Mechanical Cues.Cells. 2024 Oct 1;13(19):1638. doi: 10.3390/cells13191638. Cells. 2024. PMID: 39404401 Free PMC article. Review.
-
Cell type specification and diversity in subpallial organoids.Front Genet. 2024 Sep 26;15:1440583. doi: 10.3389/fgene.2024.1440583. eCollection 2024. Front Genet. 2024. PMID: 39391063 Free PMC article. Review.
-
Modeling Parkinson's disease pathology in human dopaminergic neurons by sequential exposure to α-synuclein fibrils and proinflammatory cytokines.Nat Neurosci. 2024 Oct 8. doi: 10.1038/s41593-024-01775-4. Online ahead of print. Nat Neurosci. 2024. PMID: 39379564
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

