Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense
- PMID: 28347402
- PMCID: PMC5370187
- DOI: 10.7554/eLife.21465
Chlamydia trachomatis-containing vacuole serves as deubiquitination platform to stabilize Mcl-1 and to interfere with host defense
Abstract
Obligate intracellular Chlamydia trachomatis replicate in a membrane-bound vacuole called inclusion, which serves as a signaling interface with the host cell. Here, we show that the chlamydial deubiquitinating enzyme (Cdu) 1 localizes in the inclusion membrane and faces the cytosol with the active deubiquitinating enzyme domain. The structure of this domain revealed high similarity to mammalian deubiquitinases with a unique α-helix close to the substrate-binding pocket. We identified the apoptosis regulator Mcl-1 as a target that interacts with Cdu1 and is stabilized by deubiquitination at the chlamydial inclusion. A chlamydial transposon insertion mutant in the Cdu1-encoding gene exhibited increased Mcl-1 and inclusion ubiquitination and reduced Mcl-1 stabilization. Additionally, inactivation of Cdu1 led to increased sensitivity of C. trachomatis for IFNγ and impaired infection in mice. Thus, the chlamydial inclusion serves as an enriched site for a deubiquitinating activity exerting a function in selective stabilization of host proteins and protection from host defense.
Keywords: Chlamydia trachomatis; Mcl-1; cell biology; cell-autonomous defense; deubiquitinase; infectious disease; microbiology.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures




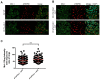
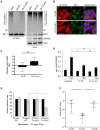
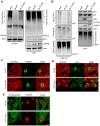
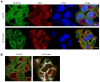

Similar articles
-
The chlamydial deubiquitinase Cdu1 supports recruitment of Golgi vesicles to the inclusion.Cell Microbiol. 2020 May;22(5):e13136. doi: 10.1111/cmi.13136. Epub 2020 Jan 20. Cell Microbiol. 2020. PMID: 31677225
-
The Human Centrosomal Protein CCDC146 Binds Chlamydia trachomatis Inclusion Membrane Protein CT288 and Is Recruited to the Periphery of the Chlamydia-Containing Vacuole.Front Cell Infect Microbiol. 2018 Jul 26;8:254. doi: 10.3389/fcimb.2018.00254. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30094225 Free PMC article.
-
Chlamydia trachomatis Is Resistant to Inclusion Ubiquitination and Associated Host Defense in Gamma Interferon-Primed Human Epithelial Cells.mBio. 2016 Dec 13;7(6):e01417-16. doi: 10.1128/mBio.01417-16. mBio. 2016. PMID: 27965446 Free PMC article.
-
Got mutants? How advances in chlamydial genetics have furthered the study of effector proteins.Pathog Dis. 2021 Feb 4;79(2):ftaa078. doi: 10.1093/femspd/ftaa078. Pathog Dis. 2021. PMID: 33512479 Free PMC article. Review.
-
Safe haven under constant attack-The Chlamydia-containing vacuole.Cell Microbiol. 2018 Oct;20(10):e12940. doi: 10.1111/cmi.12940. Epub 2018 Sep 4. Cell Microbiol. 2018. PMID: 30101516 Review.
Cited by
-
Chlamydia pan-genomic analysis reveals balance between host adaptation and selective pressure to genome reduction.BMC Genomics. 2019 Sep 12;20(1):710. doi: 10.1186/s12864-019-6059-5. BMC Genomics. 2019. PMID: 31510914 Free PMC article.
-
The evolution of regulated cell death pathways in animals and their evasion by pathogens.Physiol Rev. 2022 Jan 1;102(1):411-454. doi: 10.1152/physrev.00002.2021. Physiol Rev. 2022. PMID: 34898294 Free PMC article. Review.
-
Ubiquitin-targeted bacterial effectors: rule breakers of the ubiquitin system.EMBO J. 2023 Sep 18;42(18):e114318. doi: 10.15252/embj.2023114318. Epub 2023 Aug 9. EMBO J. 2023. PMID: 37555693 Free PMC article. Review.
-
The Chlamydia trachomatis type III secretion substrates CT142, CT143, and CT144 are secreted into the lumen of the inclusion.PLoS One. 2017 Jun 16;12(6):e0178856. doi: 10.1371/journal.pone.0178856. eCollection 2017. PLoS One. 2017. PMID: 28622339 Free PMC article.
-
Molecular pathogenesis of Chlamydia trachomatis.Front Cell Infect Microbiol. 2023 Oct 18;13:1281823. doi: 10.3389/fcimb.2023.1281823. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 37920447 Free PMC article. Review.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
-
- Adhikary S, Marinoni F, Hock A, Hulleman E, Popov N, Beier R, Bernard S, Quarto M, Capra M, Goettig S, Kogel U, Scheffner M, Helin K, Eilers M. The ubiquitin ligase HectH9 regulates transcriptional activation by myc and is essential for tumor cell proliferation. Cell. 2005;123:409–421. doi: 10.1016/j.cell.2005.08.016. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

