The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs
- PMID: 27768895
- PMCID: PMC6055481
- DOI: 10.1016/j.cell.2016.09.048
The Structure of the Polycystic Kidney Disease Channel PKD2 in Lipid Nanodiscs
Abstract
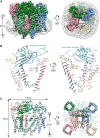
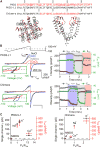
The Polycystic Kidney Disease 2 (Pkd2) gene is mutated in autosomal dominant polycystic kidney disease (ADPKD), one of the most common human monogenic disorders. Here, we present the cryo-EM structure of PKD2 in lipid bilayers at 3.0 Å resolution, which establishes PKD2 as a homotetrameric ion channel and provides insight into potential mechanisms for its activation. The PKD2 voltage-sensor domain retains two of four gating charges commonly found in those of voltage-gated ion channels. The PKD2 ion permeation pathway is constricted at the selectivity filter and near the cytoplasmic end of S6, suggesting that two gates regulate ion conduction. The extracellular domain of PKD2, a hotspot for ADPKD pathogenic mutations, contributes to channel assembly and strategically interacts with the transmembrane core, likely serving as a physical substrate for extracellular stimuli to allosterically gate the channel. Finally, our structure establishes the molecular basis for the majority of pathogenic mutations in Pkd2-related ADPKD.
Keywords: ADPKD; PKD2; TRP channel; TRPP2; cryo-EM; ion channel; polycystic kidney disease; polycystin; single particle electron cryo-microscopy.
Copyright © 2016 Elsevier Inc. All rights reserved.
Figures


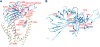

Similar articles
-
Structure of the human PKD1-PKD2 complex.Science. 2018 Sep 7;361(6406):eaat9819. doi: 10.1126/science.aat9819. Epub 2018 Aug 9. Science. 2018. PMID: 30093605
-
Structure of the polycystic kidney disease TRP channel Polycystin-2 (PC2).Nat Struct Mol Biol. 2017 Feb;24(2):114-122. doi: 10.1038/nsmb.3343. Epub 2016 Dec 19. Nat Struct Mol Biol. 2017. PMID: 27991905
-
Hydrophobic pore gates regulate ion permeation in polycystic kidney disease 2 and 2L1 channels.Nat Commun. 2018 Jun 13;9(1):2302. doi: 10.1038/s41467-018-04586-x. Nat Commun. 2018. PMID: 29899465 Free PMC article.
-
Polycystin-2 (TRPP2): Ion channel properties and regulation.Gene. 2022 Jun 15;827:146313. doi: 10.1016/j.gene.2022.146313. Epub 2022 Mar 18. Gene. 2022. PMID: 35314260 Review.
-
Vascular polycystin proteins in health and disease.Microcirculation. 2024 May;31(4):e12834. doi: 10.1111/micc.12834. Epub 2023 Oct 12. Microcirculation. 2024. PMID: 37823335 Review.
Cited by
-
Aberrant Splicing in PKD2 in a Family of Korean Patients With Autosomal Dominant Polycystic Kidney Disease.Ann Lab Med. 2024 Nov 1;44(6):621-624. doi: 10.3343/alm.2024.0221. Epub 2024 Aug 22. Ann Lab Med. 2024. PMID: 39169606 Free PMC article. No abstract available.
-
Spontaneous Coronary Artery Dissection in Patients with Autosomal Dominant Polycystic Kidney Disease: A Systematic Review of the Literature.J Pers Med. 2024 Jun 29;14(7):702. doi: 10.3390/jpm14070702. J Pers Med. 2024. PMID: 39063956 Free PMC article. Review.
-
Sensing plasma membrane pore formation induces chemokine production in survivors of regulated necrosis.Dev Cell. 2022 Jan 24;57(2):228-245.e6. doi: 10.1016/j.devcel.2021.12.015. Epub 2022 Jan 10. Dev Cell. 2022. PMID: 35016014 Free PMC article.
-
Cryo-EM in drug discovery: achievements, limitations and prospects.Nat Rev Drug Discov. 2018 Jul;17(7):471-492. doi: 10.1038/nrd.2018.77. Epub 2018 Jun 8. Nat Rev Drug Discov. 2018. PMID: 29880918 Review.
-
Identification of a single aspartate residue critical for both fast and slow calcium-dependent inactivation of the human TRPML1 channel.J Biol Chem. 2018 Jul 27;293(30):11736-11745. doi: 10.1074/jbc.RA118.003250. Epub 2018 Jun 8. J Biol Chem. 2018. PMID: 29884771 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

