Emancipating Chlamydia: Advances in the Genetic Manipulation of a Recalcitrant Intracellular Pathogen
- PMID: 27030552
- PMCID: PMC4867370
- DOI: 10.1128/MMBR.00071-15
Emancipating Chlamydia: Advances in the Genetic Manipulation of a Recalcitrant Intracellular Pathogen
Abstract
Chlamydia species infect millions of individuals worldwide and are important etiological agents of sexually transmitted disease, infertility, and blinding trachoma. Historically, the genetic intractability of this intracellular pathogen has hindered the molecular dissection of virulence factors contributing to its pathogenesis. The obligate intracellular life cycle of Chlamydia and restrictions on the use of antibiotics as selectable markers have impeded the development of molecular tools to genetically manipulate these pathogens. However, recent developments in the field have resulted in significant gains in our ability to alter the genome of Chlamydia, which will expedite the elucidation of virulence mechanisms. In this review, we discuss the challenges affecting the development of molecular genetic tools for Chlamydia and the work that laid the foundation for recent advancements in the genetic analysis of this recalcitrant pathogen.
Copyright © 2016, American Society for Microbiology. All Rights Reserved.
Figures


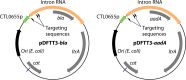
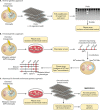
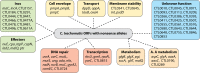
Similar articles
-
Molecular Genetic Analysis of Chlamydia Species.Annu Rev Microbiol. 2016 Sep 8;70:179-98. doi: 10.1146/annurev-micro-102215-095539. Annu Rev Microbiol. 2016. PMID: 27607551 Review.
-
Forward and Reverse Genetic Analysis of Chlamydia.Methods Mol Biol. 2019;2042:185-204. doi: 10.1007/978-1-4939-9694-0_13. Methods Mol Biol. 2019. PMID: 31385277 Free PMC article.
-
Chlamydia cell biology and pathogenesis.Nat Rev Microbiol. 2016 Jun;14(6):385-400. doi: 10.1038/nrmicro.2016.30. Epub 2016 Apr 25. Nat Rev Microbiol. 2016. PMID: 27108705 Free PMC article. Review.
-
Advances and Obstacles in the Genetic Dissection of Chlamydial Virulence.Curr Top Microbiol Immunol. 2018;412:133-158. doi: 10.1007/82_2017_76. Curr Top Microbiol Immunol. 2018. PMID: 29090367 Free PMC article. Review.
-
Advances in genetic manipulation of obligate intracellular bacterial pathogens.Front Microbiol. 2011 May 2;2:97. doi: 10.3389/fmicb.2011.00097. eCollection 2011. Front Microbiol. 2011. PMID: 21833334 Free PMC article.
Cited by
-
Development of a lambda Red based system for gene deletion in Chlamydia.PLoS One. 2024 Nov 14;19(11):e0311630. doi: 10.1371/journal.pone.0311630. eCollection 2024. PLoS One. 2024. PMID: 39541273 Free PMC article.
-
Interrogating Genes That Mediate Chlamydia trachomatis Survival in Cell Culture Using Conditional Mutants and Recombination.J Bacteriol. 2016 Jul 13;198(15):2131-9. doi: 10.1128/JB.00161-16. Print 2016 Aug 1. J Bacteriol. 2016. PMID: 27246568 Free PMC article.
-
The growing repertoire of genetic tools for dissecting chlamydial pathogenesis.Pathog Dis. 2021 May 11;79(5):ftab025. doi: 10.1093/femspd/ftab025. Pathog Dis. 2021. PMID: 33930127 Free PMC article.
-
Pathogenic Puppetry: Manipulation of the Host Actin Cytoskeleton by Chlamydia trachomatis.Int J Mol Sci. 2019 Dec 21;21(1):90. doi: 10.3390/ijms21010090. Int J Mol Sci. 2019. PMID: 31877733 Free PMC article. Review.
-
A renewed tool kit to explore Chlamydia pathogenesis: from molecular genetics to new infection models.F1000Res. 2019 Jun 21;8:F1000 Faculty Rev-935. doi: 10.12688/f1000research.18832.1. eCollection 2019. F1000Res. 2019. PMID: 31249676 Free PMC article. Review.
References
-
- Harris SR, Clarke IN, Seth-Smith HM, Solomon AW, Cutcliffe LT, Marsh P, Skilton RJ, Holland MJ, Mabey D, Peeling RW, Lewis DA, Spratt BG, Unemo M, Persson K, Bjartling C, Brunham R, de Vries HJ, Morre SA, Speksnijder A, Bebear CM, Clerc M, de Barbeyrac B, Parkhill J, Thomson NR. 2012. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet 44:413–419. doi:10.1038/ng.2214. - DOI - PMC - PubMed
-
- Peipert JF. 2003. Clinical practice. Genital chlamydial infections. N Engl J Med 349:2424–2430. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

