The Histone Demethylase KDM5 Activates Gene Expression by Recognizing Chromatin Context through Its PHD Reader Motif
- PMID: 26673323
- PMCID: PMC4684901
- DOI: 10.1016/j.celrep.2015.11.007
The Histone Demethylase KDM5 Activates Gene Expression by Recognizing Chromatin Context through Its PHD Reader Motif
Abstract
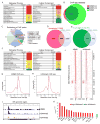
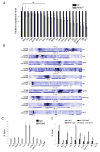
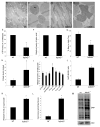
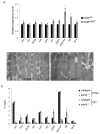
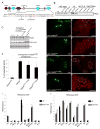
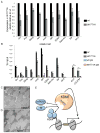
KDM5 family proteins are critically important transcriptional regulators whose physiological functions in the context of a whole animal remain largely unknown. Using genome-wide gene expression and binding analyses in Drosophila adults, we demonstrate that KDM5 (Lid) is a direct regulator of genes required for mitochondrial structure and function. Significantly, this occurs independently of KDM5's well-described JmjC domain-encoded histone demethylase activity. Instead, it requires the PHD motif of KDM5 that binds to histone H3 that is di- or trimethylated on lysine 4 (H3K4me2/3). Genome-wide, KDM5 binding overlaps with the active chromatin mark H3K4me3, and a fly strain specifically lacking H3K4me2/3 binding shows defective KDM5 promoter recruitment and gene activation. KDM5 therefore plays a central role in regulating mitochondrial function by utilizing its ability to recognize specific chromatin contexts. Importantly, KDM5-mediated regulation of mitochondrial activity is likely to be key in human diseases caused by dysfunction of this family of proteins.
Keywords: H3K4me3; KDM5; Lid; PHD motif; histone; mitochondria; transcription.
Copyright © 2015 The Authors. Published by Elsevier Inc. All rights reserved.
Figures
Similar articles
-
A Drosophila Model of Intellectual Disability Caused by Mutations in the Histone Demethylase KDM5.Cell Rep. 2018 Feb 27;22(9):2359-2369. doi: 10.1016/j.celrep.2018.02.018. Cell Rep. 2018. PMID: 29490272 Free PMC article.
-
The Histone Demethylase KDM5 Is Essential for Larval Growth in Drosophila.Genetics. 2018 Jul;209(3):773-787. doi: 10.1534/genetics.118.301004. Epub 2018 May 15. Genetics. 2018. PMID: 29764901 Free PMC article.
-
KDM5-mediated activation of genes required for mitochondrial biology is necessary for viability in Drosophila.Development. 2023 Nov 1;150(21):dev202024. doi: 10.1242/dev.202024. Epub 2023 Nov 6. Development. 2023. PMID: 37800333 Free PMC article.
-
Functions and Interactions of Mammalian KDM5 Demethylases.Front Genet. 2022 Jul 11;13:906662. doi: 10.3389/fgene.2022.906662. eCollection 2022. Front Genet. 2022. PMID: 35899196 Free PMC article. Review.
-
Diverse Functions of KDM5 in Cancer: Transcriptional Repressor or Activator?Cancers (Basel). 2022 Jul 4;14(13):3270. doi: 10.3390/cancers14133270. Cancers (Basel). 2022. PMID: 35805040 Free PMC article. Review.
Cited by
-
Proximity labeling reveals a new in vivo network of interactors for the histone demethylase KDM5.Epigenetics Chromatin. 2023 Feb 18;16(1):8. doi: 10.1186/s13072-023-00481-y. Epigenetics Chromatin. 2023. PMID: 36803422 Free PMC article.
-
BRWD3 promotes KDM5 degradation to maintain H3K4 methylation levels.Proc Natl Acad Sci U S A. 2023 Sep 26;120(39):e2305092120. doi: 10.1073/pnas.2305092120. Epub 2023 Sep 18. Proc Natl Acad Sci U S A. 2023. PMID: 37722046 Free PMC article.
-
A Drosophila Model of Intellectual Disability Caused by Mutations in the Histone Demethylase KDM5.Cell Rep. 2018 Feb 27;22(9):2359-2369. doi: 10.1016/j.celrep.2018.02.018. Cell Rep. 2018. PMID: 29490272 Free PMC article.
-
The Lid/KDM5 histone demethylase complex activates a critical effector of the oocyte-to-zygote transition.PLoS Genet. 2020 Mar 5;16(3):e1008543. doi: 10.1371/journal.pgen.1008543. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32134927 Free PMC article.
-
KDM5-mediated transcriptional activation of ribosomal protein genes alters translation efficiency to regulate mitochondrial metabolism in neurons.Nucleic Acids Res. 2024 Jun 24;52(11):6201-6219. doi: 10.1093/nar/gkae261. Nucleic Acids Res. 2024. PMID: 38597673 Free PMC article.
References
-
- Barrett A, Santangelo S, Tan K, Catchpole S, Roberts K, Spencer-Dene B, Hall D, Scibetta A, Burchell J, Verdin E, et al. Breast cancer associated transcriptional repressor PLU-1/JARID1B interacts directly with histone deacetylases. Int J Cancer. 2007;121:265–275. - PubMed
-
- Benevolenskaya EV. Histone H3K4 demethylases are essential in development and differentiation. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire. 2007a;85:435–443. - PubMed
-
- Benevolenskaya EV. Retinoblastoma binding protein 2 (RBP2) and differentiation. Biochemistry and Cell Biology-Biochimie Et Biologie Cellulaire. 2007b;85:523–523. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

