Global Mapping of the Inc-Human Interactome Reveals that Retromer Restricts Chlamydia Infection
- PMID: 26118995
- PMCID: PMC4540348
- DOI: 10.1016/j.chom.2015.06.004
Global Mapping of the Inc-Human Interactome Reveals that Retromer Restricts Chlamydia Infection
Abstract
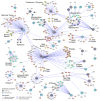
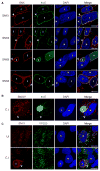
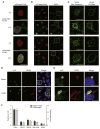
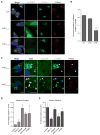
Chlamydia trachomatis is a leading cause of genital and ocular infections for which no vaccine exists. Upon entry into host cells, C. trachomatis resides within a membrane-bound compartment—the inclusion—and secretes inclusion membrane proteins (Incs) that are thought to modulate the host-bacterium interface. To expand our understanding of Inc function(s), we subjected putative C. trachomatis Incs to affinity purification-mass spectroscopy (AP-MS). We identified Inc-human interactions for 38/58 Incs with enrichment in host processes consistent with Chlamydia's intracellular life cycle. There is significant overlap between Inc targets and viral proteins, suggesting common pathogenic mechanisms among obligate intracellular microbes. IncE binds to sorting nexins (SNXs) 5/6, components of the retromer, which relocalizes SNX5/6 to the inclusion membrane and augments inclusion membrane tubulation. Depletion of retromer components enhances progeny production, revealing that retromer restricts Chlamydia infection. This study demonstrates the value of proteomics in unveiling host-pathogen interactions in genetically challenging microbes.
Copyright © 2015 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
A meta-analysis of affinity purification-mass spectrometry experimental systems used to identify eukaryotic and chlamydial proteins at the Chlamydia trachomatis inclusion membrane.J Proteomics. 2020 Feb 10;212:103595. doi: 10.1016/j.jprot.2019.103595. Epub 2019 Nov 21. J Proteomics. 2020. PMID: 31760040 Free PMC article.
-
Chlamydia interfere with an interaction between the mannose-6-phosphate receptor and sorting nexins to counteract host restriction.Elife. 2017 Mar 2;6:e22709. doi: 10.7554/eLife.22709. Elife. 2017. PMID: 28252385 Free PMC article.
-
Chlamydia trachomatis and its interaction with the cellular retromer.Int J Med Microbiol. 2018 Jan;308(1):197-205. doi: 10.1016/j.ijmm.2017.10.006. Epub 2017 Oct 26. Int J Med Microbiol. 2018. PMID: 29122514 Review.
-
The Proteome of the Isolated Chlamydia trachomatis Containing Vacuole Reveals a Complex Trafficking Platform Enriched for Retromer Components.PLoS Pathog. 2015 Jun 4;11(6):e1004883. doi: 10.1371/journal.ppat.1004883. eCollection 2015 Jun. PLoS Pathog. 2015. PMID: 26042774 Free PMC article.
-
Safe haven under constant attack-The Chlamydia-containing vacuole.Cell Microbiol. 2018 Oct;20(10):e12940. doi: 10.1111/cmi.12940. Epub 2018 Sep 4. Cell Microbiol. 2018. PMID: 30101516 Review.
Cited by
-
The Human Centrosomal Protein CCDC146 Binds Chlamydia trachomatis Inclusion Membrane Protein CT288 and Is Recruited to the Periphery of the Chlamydia-Containing Vacuole.Front Cell Infect Microbiol. 2018 Jul 26;8:254. doi: 10.3389/fcimb.2018.00254. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30094225 Free PMC article.
-
The Chlamydia trachomatis IncM Protein Interferes with Host Cell Cytokinesis, Centrosome Positioning, and Golgi Distribution and Contributes to the Stability of the Pathogen-Containing Vacuole.Infect Immun. 2023 Apr 18;91(4):e0040522. doi: 10.1128/iai.00405-22. Epub 2023 Mar 6. Infect Immun. 2023. PMID: 36877064 Free PMC article.
-
A bacterial membrane sculpting protein with BAR domain-like activity.Elife. 2021 Oct 13;10:e60049. doi: 10.7554/eLife.60049. Elife. 2021. PMID: 34643180 Free PMC article.
-
A Co-infection Model System and the Use of Chimeric Proteins to Study Chlamydia Inclusion Proteins Interaction.Front Cell Infect Microbiol. 2017 Mar 14;7:79. doi: 10.3389/fcimb.2017.00079. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28352612 Free PMC article.
-
Structural and functional insights into sorting nexin 5/6 interaction with bacterial effector IncE.Signal Transduct Target Ther. 2017 Jun 30;2:17030. doi: 10.1038/sigtrans.2017.30. eCollection 2017. Signal Transduct Target Ther. 2017. PMID: 29263922 Free PMC article.
References
-
- Bannantine JP, Griffiths RS, Viratyosin W, Brown WJ, Rockey DD. A secondary structure motif predictive of protein localization to the chlamydial inclusion membrane. Cell Microbiol. 2000;2:35–47. - PubMed
-
- Bauer MF, Hofmann S, Neupert W, Brunner M. Protein translocation into mitochondria: the role of TIM complexes. Trends in cell biology. 2000;10:25–31. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 AI081680/AI/NIAID NIH HHS/United States
- R21 AI062768/AI/NIAID NIH HHS/United States
- A121844/PHS HHS/United States
- T32 AI007334/AI/NIAID NIH HHS/United States
- R21 AI105561/AI/NIAID NIH HHS/United States
- R01 AI100759/AI/NIAID NIH HHS/United States
- R01AI101441/AI/NIAID NIH HHS/United States
- P01 091575/PHS HHS/United States
- P50 GM081879/GM/NIGMS NIH HHS/United States
- K08 AI091656/AI/NIAID NIH HHS/United States
- R01 AI073770/AI/NIAID NIH HHS/United States
- P01 090935/PHS HHS/United States
- AI081694/AI/NIAID NIH HHS/United States
- K08 AI104766/AI/NIAID NIH HHS/United States
- P50 GM082250/GM/NIGMS NIH HHS/United States
- AI100759/AI/NIAID NIH HHS/United States
- P0065678/PHS HHS/United States
- K08AI091656/AI/NIAID NIH HHS/United States
- P30 AI027763/AI/NIAID NIH HHS/United States
- R01 AI101441/AI/NIAID NIH HHS/United States
- R01 AI039558/AI/NIAID NIH HHS/United States
- U19 AI106754/AI/NIAID NIH HHS/United States
- U19 AI113187/AI/NIAID NIH HHS/United States
- U54AI081680/AI/NIAID NIH HHS/United States
- R01 AI081694/AI/NIAID NIH HHS/United States
- AI105561/AI/NIAID NIH HHS/United States
- CA-0063244/CA/NCI NIH HHS/United States
- P01 AI090935/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

