Genes from scratch--the evolutionary fate of de novo genes
- PMID: 25773713
- PMCID: PMC4383367
- DOI: 10.1016/j.tig.2015.02.007
Genes from scratch--the evolutionary fate of de novo genes
Abstract
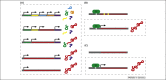
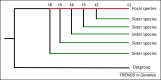
Although considered an extremely unlikely event, many genes emerge from previously noncoding genomic regions. This review covers the entire life cycle of such de novo genes. Two competing hypotheses about the process of de novo gene birth are discussed as well as the high death rate of de novo genes. Despite the high death rate, some de novo genes are retained and remain functional, even in distantly related species, through their integration into gene networks. Further studies combining gene expression with ribosome profiling in multiple populations across different species will be instrumental for an improved understanding of the evolutionary processes operating on de novo genes.
Keywords: de novo genes; orphans; population genetics; transcription.
Copyright © 2015 The Author. Published by Elsevier Ltd.. All rights reserved.
Figures
Similar articles
-
Origins, evolution, and physiological implications of de novo genes in yeast.Yeast. 2022 Sep;39(9):471-481. doi: 10.1002/yea.3810. Epub 2022 Aug 24. Yeast. 2022. PMID: 35959631 Free PMC article. Review.
-
Tracing the De Novo Origin of Protein-Coding Genes in Yeast.mBio. 2018 Jul 31;9(4):e01024-18. doi: 10.1128/mBio.01024-18. mBio. 2018. PMID: 30065088 Free PMC article.
-
Four classic "de novo" genes all have plausible homologs and likely evolved from retro-duplicated or pseudogenic sequences.Mol Genet Genomics. 2024 Feb 5;299(1):6. doi: 10.1007/s00438-023-02090-6. Mol Genet Genomics. 2024. PMID: 38315248
-
Protein evidence of unannotated ORFs in Drosophila reveals diversity in the evolution and properties of young proteins.Elife. 2022 Sep 30;11:e78772. doi: 10.7554/eLife.78772. Elife. 2022. PMID: 36178469 Free PMC article.
-
Orphans and new gene origination, a structural and evolutionary perspective.Curr Opin Struct Biol. 2014 Jun;26:73-83. doi: 10.1016/j.sbi.2014.05.006. Epub 2014 Jun 13. Curr Opin Struct Biol. 2014. PMID: 24934869 Review.
Cited by
-
The draft genome of Actinia tenebrosa reveals insights into toxin evolution.Ecol Evol. 2019 Sep 18;9(19):11314-11328. doi: 10.1002/ece3.5633. eCollection 2019 Oct. Ecol Evol. 2019. PMID: 31641475 Free PMC article.
-
A hotspot for new genes.Elife. 2019 Aug 29;8:e50136. doi: 10.7554/eLife.50136. Elife. 2019. PMID: 31464682 Free PMC article.
-
Origins, evolution, and physiological implications of de novo genes in yeast.Yeast. 2022 Sep;39(9):471-481. doi: 10.1002/yea.3810. Epub 2022 Aug 24. Yeast. 2022. PMID: 35959631 Free PMC article. Review.
-
New Genes and Functional Innovation in Mammals.Genome Biol Evol. 2017 Jul 1;9(7):1886-1900. doi: 10.1093/gbe/evx136. Genome Biol Evol. 2017. PMID: 28854603 Free PMC article.
-
The brachyceran de novo gene PIP82, a phosphorylation target of aPKC, is essential for proper formation and maintenance of the rhabdomeric photoreceptor apical domain in Drosophila.PLoS Genet. 2020 Jun 24;16(6):e1008890. doi: 10.1371/journal.pgen.1008890. eCollection 2020 Jun. PLoS Genet. 2020. PMID: 32579558 Free PMC article.
References
-
- Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. - PubMed
-
- Dujon B. The yeast genome project: what did we learn? Trends Genet. 1996;12:263–270. - PubMed
-
- Fischer D., Eisenberg D. Finding families for genomic ORFans. Bioinformatics. 1999;15:759–762. - PubMed
-
- Tautz D., Domazet-Loso T. The evolutionary origin of orphan genes. Nat. Rev. Genet. 2011;12:692–702. - PubMed
-
- Khalturin K. More than just orphans: are taxonomically-restricted genes important in evolution? Trends Genet. 2009;25:404–413. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

