Rapid, specific, no-wash, far-red fluorogen activation in subcellular compartments by targeted fluorogen activating proteins
- PMID: 25650487
- PMCID: PMC4867890
- DOI: 10.1021/cb500957k
Rapid, specific, no-wash, far-red fluorogen activation in subcellular compartments by targeted fluorogen activating proteins
Abstract
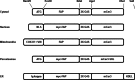
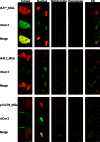
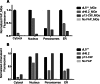
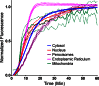
Live cell imaging requires bright photostable dyes that can target intracellular organelles and proteins with high specificity in a no-wash protocol. Organic dyes possess the desired photochemical properties and can be covalently linked to various protein tags. The currently available fluorogenic dyes are in the green/yellow range where there is high cellular autofluorescence and the near-infrared (NIR) dyes need to be washed out. Protein-mediated activation of far-red fluorogenic dyes has the potential to address these challenges because the cell-permeant dye is small and nonfluorescent until bound to its activating protein, and this binding is rapid. In this study, three single chain variable fragment (scFv)-derived fluorogen activating proteins (FAPs), which activate far-red emitting fluorogens, were evaluated for targeting, brightness, and photostability in the cytosol, nucleus, mitochondria, peroxisomes, and endoplasmic reticulum with a cell-permeant malachite green analog in cultured mammalian cells. Efficient labeling was achieved within 20-30 min for each protein upon the addition of nM concentrations of dye, producing a signal that colocalized significantly with a linked mCerulean3 (mCer3) fluorescent protein and organelle specific dyes but showed divergent photostability and brightness properties dependent on the FAP. These FAPs and the ester of malachite green dye (MGe) can be used as specific, rapid, and wash-free labels for intracellular sites in live cells with far-red excitation and emission properties, useful in a variety of multicolor experiments.
Conflict of interest statement
The authors declare the following competing financial interest(s): M.B. is a founder of Sharp Edge Labs, which aims to exploit FAP-based labeling for applications in drug discovery.
Figures
Similar articles
-
Fluorogen-activating single-chain antibodies for imaging cell surface proteins.Nat Biotechnol. 2008 Feb;26(2):235-40. doi: 10.1038/nbt1368. Epub 2007 Dec 23. Nat Biotechnol. 2008. PMID: 18157118
-
Multiexcitation Fluorogenic Labeling of Surface, Intracellular, and Total Protein Pools in Living Cells.Bioconjug Chem. 2016 Jun 15;27(6):1525-31. doi: 10.1021/acs.bioconjchem.6b00169. Epub 2016 May 19. Bioconjug Chem. 2016. PMID: 27159569 Free PMC article.
-
Generation of Fluorogen-Activating Designed Ankyrin Repeat Proteins (FADAs) as Versatile Sensor Tools.J Mol Biol. 2016 Mar 27;428(6):1272-1289. doi: 10.1016/j.jmb.2016.01.017. Epub 2016 Jan 23. J Mol Biol. 2016. PMID: 26812208
-
Fluorogen-Activating Proteins: Next-Generation Fluorescence Probes for Biological Research.Bioconjug Chem. 2020 Jan 15;31(1):16-27. doi: 10.1021/acs.bioconjchem.9b00710. Epub 2019 Dec 13. Bioconjug Chem. 2020. PMID: 31789501 Review.
-
Fluorogen-activating proteins: beyond classical fluorescent proteins.Acta Pharm Sin B. 2018 May;8(3):339-348. doi: 10.1016/j.apsb.2018.02.001. Epub 2018 Mar 24. Acta Pharm Sin B. 2018. PMID: 29881673 Free PMC article. Review.
Cited by
-
Fluorescence-Based Quantitative Synapse Analysis for Cell Type-Specific Connectomics.eNeuro. 2019 Oct 31;6(5):ENEURO.0193-19.2019. doi: 10.1523/ENEURO.0193-19.2019. Print 2019 Sep/Oct. eNeuro. 2019. PMID: 31548370 Free PMC article.
-
Fluorogenic Photoaffinity Labeling of Proteins in Living Cells.Bioconjug Chem. 2019 May 15;30(5):1309-1313. doi: 10.1021/acs.bioconjchem.9b00203. Epub 2019 Apr 17. Bioconjug Chem. 2019. PMID: 30978287 Free PMC article.
-
Visualizing Cardiolipin In Situ with HKCL-1M, a Highly Selective and Sensitive Fluorescent Probe.J Am Chem Soc. 2023 May 24;145(20):11311-11322. doi: 10.1021/jacs.3c00243. Epub 2023 Apr 27. J Am Chem Soc. 2023. PMID: 37103240 Free PMC article.
-
Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo.Proc Natl Acad Sci U S A. 2016 Jan 19;113(3):497-502. doi: 10.1073/pnas.1513094113. Epub 2015 Dec 28. Proc Natl Acad Sci U S A. 2016. PMID: 26711992 Free PMC article.
-
Cell-cell communication: new insights and clinical implications.Signal Transduct Target Ther. 2024 Aug 7;9(1):196. doi: 10.1038/s41392-024-01888-z. Signal Transduct Target Ther. 2024. PMID: 39107318 Free PMC article. Review.
References
-
- Wiedenmann J.; Oswald F.; Nienhaus G. U. (2009) Fluorescent Proteins for Live Cell Imaging: Opportunities, Limitations, and Challenges. IUBMB Life 61, 1029–1042. - PubMed
-
- Chen S.; Chen Z. J.; Ren W.; Ai H. W. (2012) Reaction-Based Genetically Encoded Fluorescent Hydrogen Sulfide Sensors. J. Am. Chem. Soc. 134, 9589–9592. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

