Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells
- PMID: 25081482
- PMCID: PMC4459787
- DOI: 10.1126/science.1254917
Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells
Abstract
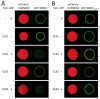
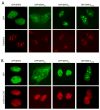
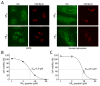
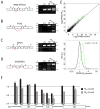
Many RNA regulatory proteins controlling pre-messenger RNA splicing contain serine:arginine (SR) repeats. Here, we found that these SR domains bound hydrogel droplets composed of fibrous polymers of the low-complexity domain of heterogeneous ribonucleoprotein A2 (hnRNPA2). Hydrogel binding was reversed upon phosphorylation of the SR domain by CDC2-like kinases 1 and 2 (CLK1/2). Mutated variants of the SR domains changing serine to glycine (SR-to-GR variants) also bound to hnRNPA2 hydrogels but were not affected by CLK1/2. When expressed in mammalian cells, these variants bound nucleoli. The translation products of the sense and antisense transcripts of the expansion repeats associated with the C9orf72 gene altered in neurodegenerative disease encode GRn and PRn repeat polypeptides. Both peptides bound to hnRNPA2 hydrogels independent of CLK1/2 activity. When applied to cultured cells, both peptides entered cells, migrated to the nucleus, bound nucleoli, and poisoned RNA biogenesis, which caused cell death.
Copyright © 2014, American Association for the Advancement of Science.
Figures


Comment in
-
Cell Biology. Clogging information flow in ALS.Science. 2014 Sep 5;345(6201):1118-9. doi: 10.1126/science.1259461. Science. 2014. PMID: 25190778 No abstract available.
Similar articles
-
Cell Biology. Clogging information flow in ALS.Science. 2014 Sep 5;345(6201):1118-9. doi: 10.1126/science.1259461. Science. 2014. PMID: 25190778 No abstract available.
-
Toxic PR Poly-Dipeptides Encoded by the C9orf72 Repeat Expansion Target LC Domain Polymers.Cell. 2016 Oct 20;167(3):789-802.e12. doi: 10.1016/j.cell.2016.10.003. Cell. 2016. PMID: 27768897 Free PMC article.
-
Poly-dipeptides encoded by the C9ORF72 repeats block global protein translation.Hum Mol Genet. 2016 May 1;25(9):1803-13. doi: 10.1093/hmg/ddw052. Epub 2016 Feb 29. Hum Mol Genet. 2016. PMID: 26931465 Free PMC article.
-
Genetic models of C9orf72: what is toxic?Curr Opin Genet Dev. 2017 Jun;44:92-101. doi: 10.1016/j.gde.2017.01.006. Epub 2017 Mar 29. Curr Opin Genet Dev. 2017. PMID: 28364657 Review.
-
Pathogenic determinants and mechanisms of ALS/FTD linked to hexanucleotide repeat expansions in the C9orf72 gene.Neurosci Lett. 2017 Jan 1;636:16-26. doi: 10.1016/j.neulet.2016.09.007. Epub 2016 Sep 13. Neurosci Lett. 2017. PMID: 27619540 Free PMC article. Review.
Cited by
-
C9orf72 arginine-rich dipeptide repeats inhibit UPF1-mediated RNA decay via translational repression.Nat Commun. 2020 Jul 3;11(1):3354. doi: 10.1038/s41467-020-17129-0. Nat Commun. 2020. PMID: 32620797 Free PMC article.
-
Identification of selective and non-selective C9ORF72 targeting in vivo active siRNAs.Mol Ther Nucleic Acids. 2024 Jul 31;35(3):102291. doi: 10.1016/j.omtn.2024.102291. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 39233852 Free PMC article.
-
The expanding syndrome of amyotrophic lateral sclerosis: a clinical and molecular odyssey.J Neurol Neurosurg Psychiatry. 2015 Jun;86(6):667-73. doi: 10.1136/jnnp-2014-308946. Epub 2015 Feb 2. J Neurol Neurosurg Psychiatry. 2015. PMID: 25644224 Free PMC article. Review.
-
Role of C9orf72 hexanucleotide repeat expansions in ALS/FTD pathogenesis.Front Mol Neurosci. 2024 Jan 22;17:1322720. doi: 10.3389/fnmol.2024.1322720. eCollection 2024. Front Mol Neurosci. 2024. PMID: 38318532 Free PMC article. Review.
-
ALS Genetics: Gains, Losses, and Implications for Future Therapies.Neuron. 2020 Dec 9;108(5):822-842. doi: 10.1016/j.neuron.2020.08.022. Epub 2020 Sep 14. Neuron. 2020. PMID: 32931756 Free PMC article. Review.
References
-
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung GY, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. - DOI - PMC - PubMed
-
- Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita VM, Kaivorinne AL, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ ITALSGEN Consortium. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. - DOI - PMC - PubMed
-
- Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li HR, Jiang J, Watt AT, Chun S, Katz M, Qiu J, Sun Y, Ling SC, Zhu Q, Polymenidou M, Drenner K, Artates JW, McAlonis-Downes M, Markmiller S, Hutt KR, Pizzo DP, Cady J, Harms MB, Baloh RH, Vandenberg SR, Yeo GW, Fu XD, Bennett CF, Cleveland DW, Ravits J. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc Natl Acad Sci USA. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. - DOI - PMC - PubMed
-
- Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, Ostrow LW, Rothstein JD, Troncoso JC, Ranum LP. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

