TRPV1 structures in distinct conformations reveal activation mechanisms
- PMID: 24305161
- PMCID: PMC4023639
- DOI: 10.1038/nature12823
TRPV1 structures in distinct conformations reveal activation mechanisms
Abstract
Transient receptor potential (TRP) channels are polymodal signal detectors that respond to a wide range of physical and chemical stimuli. Elucidating how these channels integrate and convert physiological signals into channel opening is essential to understanding how they regulate cell excitability under normal and pathophysiological conditions. Here we exploit pharmacological probes (a peptide toxin and small vanilloid agonists) to determine structures of two activated states of the capsaicin receptor, TRPV1. A domain (consisting of transmembrane segments 1-4) that moves during activation of voltage-gated channels remains stationary in TRPV1, highlighting differences in gating mechanisms for these structurally related channel superfamilies. TRPV1 opening is associated with major structural rearrangements in the outer pore, including the pore helix and selectivity filter, as well as pronounced dilation of a hydrophobic constriction at the lower gate, suggesting a dual gating mechanism. Allosteric coupling between upper and lower gates may account for rich physiological modulation exhibited by TRPV1 and other TRP channels.
Figures



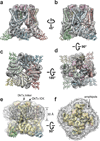
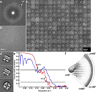
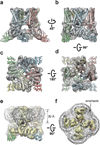
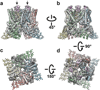
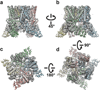
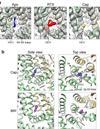
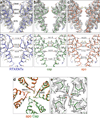
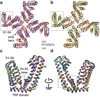
Comment in
-
Structural biology: Ion channel seen by electron microscopy.Nature. 2013 Dec 5;504(7478):93-4. doi: 10.1038/504093a. Nature. 2013. PMID: 24305155 No abstract available.
-
Multi-Functional Diarylurea Small Molecule Inhibitors of TRPV1 with Therapeutic Potential for Neuroinflammation.AAPS J. 2016 Jul;18(4):898-913. doi: 10.1208/s12248-016-9888-z. Epub 2016 Mar 21. AAPS J. 2016. PMID: 27000851 Free PMC article.
Similar articles
-
Conformational dynamics in TRPV1 channels reported by an encoded coumarin amino acid.Elife. 2017 Dec 5;6:e28626. doi: 10.7554/eLife.28626. Elife. 2017. PMID: 29206105 Free PMC article.
-
Mutation of I696 and W697 in the TRP box of vanilloid receptor subtype I modulates allosteric channel activation.J Gen Physiol. 2014 Mar;143(3):361-75. doi: 10.1085/jgp.201311070. J Gen Physiol. 2014. PMID: 24567510 Free PMC article.
-
The region adjacent to the C-end of the inner gate in transient receptor potential melastatin 8 (TRPM8) channels plays a central role in allosteric channel activation.J Biol Chem. 2014 Oct 10;289(41):28579-94. doi: 10.1074/jbc.M114.577478. Epub 2014 Aug 25. J Biol Chem. 2014. PMID: 25157108 Free PMC article.
-
ThermoTRP channels as modular proteins with allosteric gating.Cell Calcium. 2007 Oct-Nov;42(4-5):427-38. doi: 10.1016/j.ceca.2007.04.004. Epub 2007 May 17. Cell Calcium. 2007. PMID: 17499848 Review.
-
Polymodal Transient Receptor Potential Vanilloid Type 1 Nocisensor: Structure, Modulators, and Therapeutic Applications.Adv Protein Chem Struct Biol. 2016;104:81-125. doi: 10.1016/bs.apcsb.2015.11.005. Epub 2016 Jan 4. Adv Protein Chem Struct Biol. 2016. PMID: 27038373 Review.
Cited by
-
Evolutionary dynamics of metazoan TRP channels.Pflugers Arch. 2015 Oct;467(10):2043-53. doi: 10.1007/s00424-015-1705-5. Epub 2015 Apr 1. Pflugers Arch. 2015. PMID: 25823501 Review.
-
Capsaicin and Proton Differently Modulate Activation Kinetics of Mouse Transient Receptor Potential Vanilloid-1 Channel Induced by Depolarization.Front Pharmacol. 2021 May 20;12:672157. doi: 10.3389/fphar.2021.672157. eCollection 2021. Front Pharmacol. 2021. PMID: 34093200 Free PMC article.
-
2.8 Å resolution reconstruction of the Thermoplasma acidophilum 20S proteasome using cryo-electron microscopy.Elife. 2015 Mar 11;4:e06380. doi: 10.7554/eLife.06380. Elife. 2015. PMID: 25760083 Free PMC article.
-
Exploring functional roles of TRPV1 intracellular domains with unstructured peptide-insertion screening.Sci Rep. 2016 Sep 26;6:33827. doi: 10.1038/srep33827. Sci Rep. 2016. PMID: 27666400 Free PMC article.
-
TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action.Nature. 2016 Jun 16;534(7607):347-51. doi: 10.1038/nature17964. Epub 2016 May 18. Nature. 2016. PMID: 27281200 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

