LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
- PMID: 23260659
- PMCID: PMC3625064
- DOI: 10.1016/j.molcel.2012.11.019
LSD2/KDM1B and its cofactor NPAC/GLYR1 endow a structural and molecular model for regulation of H3K4 demethylation
Abstract
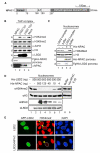
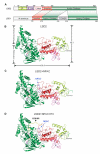
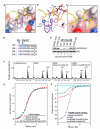
Dynamic regulation of histone methylation represents a fundamental epigenetic mechanism underlying eukaryotic gene regulation, yet little is known about how the catalytic activities of histone demethylases are regulated. Here, we identify and characterize NPAC/GLYR1 as an LSD2/KDM1b-specific cofactor that stimulates H3K4me1 and H3K4me2 demethylation. We determine the crystal structures of LSD2 alone and LSD2 in complex with the NPAC linker region in the absence or presence of histone H3 peptide, at resolutions of 2.9, 2.0, and 2.25 Å, respectively. These crystal structures and further biochemical characterization define a dodecapeptide of NPAC (residues 214-225) as the minimal functional unit for its cofactor activity and provide structural determinants and a molecular mechanism underlying the intrinsic cofactor activity of NPAC in stimulating LSD2-catalyzed H3K4 demethylation. Thus, these findings establish a model for how a cofactor directly regulates histone demethylation and will have a significant impact on our understanding of catalytic-activity-based epigenetic regulation.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
The NPAC-LSD2 complex in nucleosome demethylation.Enzymes. 2023;53:97-111. doi: 10.1016/bs.enz.2023.03.003. Epub 2023 Apr 21. Enzymes. 2023. PMID: 37748839 Review.
-
Evaluation of phenylcyclopropylamine compounds by enzymatic assay of lysine-specific demethylase 2 in the presence of NPAC peptide.Bioorg Med Chem Lett. 2016 Feb 15;26(4):1193-5. doi: 10.1016/j.bmcl.2016.01.036. Epub 2016 Jan 16. Bioorg Med Chem Lett. 2016. PMID: 26794039
-
Structure-function analysis reveals a novel mechanism for regulation of histone demethylase LSD2/AOF1/KDM1b.Cell Res. 2013 Feb;23(2):225-41. doi: 10.1038/cr.2012.177. Epub 2012 Dec 25. Cell Res. 2013. PMID: 23266887 Free PMC article.
-
A Tail-Based Mechanism Drives Nucleosome Demethylation by the LSD2/NPAC Multimeric Complex.Cell Rep. 2019 Apr 9;27(2):387-399.e7. doi: 10.1016/j.celrep.2019.03.061. Cell Rep. 2019. PMID: 30970244
-
Regulation of histone methylation by demethylimination and demethylation.Nat Rev Mol Cell Biol. 2007 Apr;8(4):307-18. doi: 10.1038/nrm2143. Epub 2007 Mar 7. Nat Rev Mol Cell Biol. 2007. PMID: 17342184 Review.
Cited by
-
Targeting KDM1B-dependent miR-215-AR-AGR2-axis promotes sensitivity to enzalutamide-resistant prostate cancer.Cancer Gene Ther. 2022 May;29(5):543-557. doi: 10.1038/s41417-021-00332-6. Epub 2021 Apr 14. Cancer Gene Ther. 2022. Retraction in: Cancer Gene Ther. 2023 Oct;30(10):1442. doi: 10.1038/s41417-023-00669-0 PMID: 33854217 Retracted.
-
Structure of a Naegleria Tet-like dioxygenase in complex with 5-methylcytosine DNA.Nature. 2014 Feb 20;506(7488):391-5. doi: 10.1038/nature12905. Epub 2013 Dec 25. Nature. 2014. PMID: 24390346 Free PMC article.
-
Structure and function of dioxygenases in histone demethylation and DNA/RNA demethylation.IUCrJ. 2014 Oct 28;1(Pt 6):540-9. doi: 10.1107/S2052252514020922. eCollection 2014 Nov 1. IUCrJ. 2014. PMID: 25485134 Free PMC article. Review.
-
Interplay among nucleosomal DNA, histone tails, and corepressor CoREST underlies LSD1-mediated H3 demethylation.Proc Natl Acad Sci U S A. 2015 Mar 3;112(9):2752-7. doi: 10.1073/pnas.1419468112. Epub 2015 Feb 17. Proc Natl Acad Sci U S A. 2015. PMID: 25730864 Free PMC article.
-
NDF, a nucleosome-destabilizing factor that facilitates transcription through nucleosomes.Genes Dev. 2018 May 1;32(9-10):682-694. doi: 10.1101/gad.313973.118. Epub 2018 May 14. Genes Dev. 2018. PMID: 29759984 Free PMC article.
References
-
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. - PubMed
-
- Allis CD, Berger SL, Cote J, Dent S, Jenuwien T, Kouzarides T, Pillus L, Reinberg D, Shi Y, Shiekhattar R, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–636. - PubMed
-
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. - PubMed
-
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 2007;14:1008–1016. - PubMed
-
- Binda C, Valente S, Romanenghi M, Pilotto S, Cirilli R, Karytinos A, Ciossani G, Botrugno OA, Forneris F, Tardugno M, et al. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J. Am. Chem. Soc. 2010;132:6827–6833. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

