Conditional control of gene function by an invertible gene trap in zebrafish
- PMID: 22908272
- PMCID: PMC3458342
- DOI: 10.1073/pnas.1206131109
Conditional control of gene function by an invertible gene trap in zebrafish
Abstract
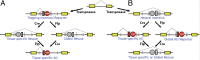
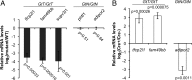
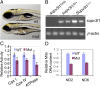
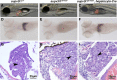
Conditional mutations are essential for determining the stage- and tissue-specific functions of genes. Here we achieve conditional mutagenesis in zebrafish using FT1, a gene-trap cassette that can be stably inverted by both Cre and Flp recombinases. We demonstrate that intronic insertions in the gene-trapping orientation severely disrupt the expression of the host gene, whereas intronic insertions in the neutral orientation do not significantly affect host gene expression. Cre- and Flp-mediated recombination switches the orientation of the gene-trap cassette, permitting conditional rescue in one orientation and conditional knockout in the other. To illustrate the utility of this system we analyzed the functional consequence of intronic FT1 insertion in supv3l1, a gene encoding a mitochondrial RNA helicase. Global supv311 mutants have impaired mitochondrial function, embryonic lethality, and agenesis of the liver. Conditional rescue of supv311 expression in hepatocytes specifically corrected the liver defects. To test whether the liver function of supv311 is required for viability we used Flp-mediated recombination in the germline to generate a neutral allele at the locus. Subsequently, tissue-specific expression of Cre conditionally inactivated the targeted locus. Hepatocyte-specific inactivation of supv311 caused liver degeneration, growth retardation, and juvenile lethality, a phenotype that was less severe than the global disruption of supv311. Thus, supv311 is required in multiple tissues for organismal viability. Our mutagenesis approach is very efficient and could be used to generate conditional alleles throughout the zebrafish genome. Furthermore, because FT1 is based on the promiscuous Tol2 transposon, it should be applicable to many organisms.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
The changing conditions of zebrafish mutants.Proc Natl Acad Sci U S A. 2012 Sep 18;109(38):15082-3. doi: 10.1073/pnas.1212832109. Epub 2012 Sep 4. Proc Natl Acad Sci U S A. 2012. PMID: 22949684 Free PMC article. No abstract available.
Similar articles
-
Cre/lox regulated conditional rescue and inactivation with zebrafish UFlip alleles generated by CRISPR-Cas9 targeted integration.Elife. 2022 Jun 17;11:e71478. doi: 10.7554/eLife.71478. Elife. 2022. PMID: 35713402 Free PMC article.
-
Generation of a conditional lima1a allele in zebrafish using the FLEx switch technology.Genesis. 2016 Jan;54(1):19-28. doi: 10.1002/dvg.22909. Epub 2015 Dec 6. Genesis. 2016. PMID: 26572123
-
Conditional gene-trap mutagenesis in zebrafish.Methods Mol Biol. 2014;1101:393-411. doi: 10.1007/978-1-62703-721-1_19. Methods Mol Biol. 2014. PMID: 24233792 Free PMC article.
-
Mammalian genome targeting using site-specific recombinases.Front Biosci. 2006 Jan 1;11:1108-36. doi: 10.2741/1867. Front Biosci. 2006. PMID: 16146801 Review.
-
Conditional somatic mutagenesis in the mouse using site-specific recombinases.Handb Exp Pharmacol. 2007;(178):3-28. doi: 10.1007/978-3-540-35109-2_1. Handb Exp Pharmacol. 2007. PMID: 17203649 Review.
Cited by
-
A HIT-trapping strategy for rapid generation of reversible and conditional alleles using a universal donor.Genome Res. 2021 May;31(5):900-909. doi: 10.1101/gr.271312.120. Epub 2021 Apr 1. Genome Res. 2021. PMID: 33795333 Free PMC article.
-
Building the vertebrate codex using the gene breaking protein trap library.Elife. 2020 Aug 11;9:e54572. doi: 10.7554/eLife.54572. Elife. 2020. PMID: 32779569 Free PMC article.
-
The role of retrograde intraflagellar transport genes in aminoglycoside-induced hair cell death.Biol Open. 2019 Jan 14;8(1):bio038745. doi: 10.1242/bio.038745. Biol Open. 2019. PMID: 30578252 Free PMC article.
-
Cre/lox regulated conditional rescue and inactivation with zebrafish UFlip alleles generated by CRISPR-Cas9 targeted integration.Elife. 2022 Jun 17;11:e71478. doi: 10.7554/eLife.71478. Elife. 2022. PMID: 35713402 Free PMC article.
-
Efficient disruption of Zebrafish genes using a Gal4-containing gene trap.BMC Genomics. 2013 Sep 14;14:619. doi: 10.1186/1471-2164-14-619. BMC Genomics. 2013. PMID: 24034702 Free PMC article.
References
-
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science. 1994;265:103–106. - PubMed
-
- Gossler A, Joyner AL, Rossant J, Skarnes WC. Mouse embryonic stem cells and reporter constructs to detect developmentally regulated genes. Science. 1989;244:463–465. - PubMed
-
- Schnütgen F, et al. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

