Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel
- PMID: 22678295
- PMCID: PMC3979295
- DOI: 10.1038/nature11054
Crystal structure of an orthologue of the NaChBac voltage-gated sodium channel
Abstract
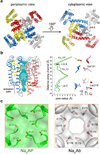
Voltage-gated sodium (Na(v)) channels are essential for the rapid depolarization of nerve and muscle, and are important drug targets. Determination of the structures of Na(v) channels will shed light on ion channel mechanisms and facilitate potential clinical applications. A family of bacterial Na(v) channels, exemplified by the Na(+)-selective channel of bacteria (NaChBac), provides a useful model system for structure-function analysis. Here we report the crystal structure of Na(v)Rh, a NaChBac orthologue from the marine alphaproteobacterium HIMB114 (Rickettsiales sp. HIMB114; denoted Rh), at 3.05 Å resolution. The channel comprises an asymmetric tetramer. The carbonyl oxygen atoms of Thr 178 and Leu 179 constitute an inner site within the selectivity filter where a hydrated Ca(2+) resides in the crystal structure. The outer mouth of the Na(+) selectivity filter, defined by Ser 181 and Glu 183, is closed, as is the activation gate at the intracellular side of the pore. The voltage sensors adopt a depolarized conformation in which all the gating charges are exposed to the extracellular environment. We propose that Na(v)Rh is in an 'inactivated' conformation. Comparison of Na(v)Rh with Na(v)Ab reveals considerable conformational rearrangements that may underlie the electromechanical coupling mechanism of voltage-gated channels.
Figures

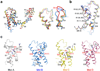

Similar articles
-
Models of the structure and gating mechanisms of the pore domain of the NaChBac ion channel.Biophys J. 2008 Oct;95(8):3650-62. doi: 10.1529/biophysj.108.135327. Epub 2008 Jul 18. Biophys J. 2008. PMID: 18641075 Free PMC article.
-
Crystal structure of a voltage-gated sodium channel in two potentially inactivated states.Nature. 2012 May 20;486(7401):135-9. doi: 10.1038/nature11077. Nature. 2012. PMID: 22678296 Free PMC article.
-
The crystal structure of a voltage-gated sodium channel.Nature. 2011 Jul 10;475(7356):353-8. doi: 10.1038/nature10238. Nature. 2011. PMID: 21743477 Free PMC article.
-
Mechanisms of Drug Binding to Voltage-Gated Sodium Channels.Handb Exp Pharmacol. 2018;246:209-231. doi: 10.1007/164_2017_73. Handb Exp Pharmacol. 2018. PMID: 29138928 Review.
-
Insect-selective spider toxins targeting voltage-gated sodium channels.Toxicon. 2007 Mar 15;49(4):490-512. doi: 10.1016/j.toxicon.2006.11.027. Epub 2006 Dec 5. Toxicon. 2007. PMID: 17223149 Review.
Cited by
-
Na(+)/Ca(2+) selectivity in the bacterial voltage-gated sodium channel NavAb.PeerJ. 2013 Feb 12;1:e16. doi: 10.7717/peerj.16. Print 2013. PeerJ. 2013. PMID: 23638350 Free PMC article.
-
Brevetoxin and Conotoxin Interactions with Single-Domain Voltage-Gated Sodium Channels from a Diatom and Coccolithophore.Mar Drugs. 2021 Mar 2;19(3):140. doi: 10.3390/md19030140. Mar Drugs. 2021. PMID: 33801270 Free PMC article.
-
Exploring volatile general anesthetic binding to a closed membrane-bound bacterial voltage-gated sodium channel via computation.PLoS Comput Biol. 2013;9(6):e1003090. doi: 10.1371/journal.pcbi.1003090. Epub 2013 Jun 13. PLoS Comput Biol. 2013. PMID: 23785267 Free PMC article.
-
Breaking the Backbone: Central Arginine Residues Induce Membrane Exit and Helix Distortions within a Dynamic Membrane Peptide.J Phys Chem B. 2019 Sep 26;123(38):8034-8047. doi: 10.1021/acs.jpcb.9b06034. Epub 2019 Sep 17. J Phys Chem B. 2019. PMID: 31483653 Free PMC article.
-
Propofol inhibits the voltage-gated sodium channel NaChBac at multiple sites.J Gen Physiol. 2018 Sep 3;150(9):1317-1331. doi: 10.1085/jgp.201811993. Epub 2018 Jul 17. J Gen Physiol. 2018. PMID: 30018039 Free PMC article.
References
-
- Hille B. Ion channels of excitable membranes. Sunderland, MA: Sinauer Associates; 2001. p. 814.
-
- Mantegazza M, Curia G, Biagini G, Ragsdale DS, Avoli M. Voltage-gated sodium channels as therapeutic targets in epilepsy and other neurological disorders. Lancet Neurol. 2010;9:413–424. - PubMed
-
- Ren D, et al. A prokaryotic voltage-gated sodium channel. Science. 2001;294:2372–2375. - PubMed
-
- Catterall WA. The molecular basis of neuronal excitability. Science. 1984;223:653–661. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

