A flexible Bayesian model for studying gene-environment interaction
- PMID: 22291610
- PMCID: PMC3266891
- DOI: 10.1371/journal.pgen.1002482
A flexible Bayesian model for studying gene-environment interaction
Abstract
An important follow-up step after genetic markers are found to be associated with a disease outcome is a more detailed analysis investigating how the implicated gene or chromosomal region and an established environment risk factor interact to influence the disease risk. The standard approach to this study of gene-environment interaction considers one genetic marker at a time and therefore could misrepresent and underestimate the genetic contribution to the joint effect when one or more functional loci, some of which might not be genotyped, exist in the region and interact with the environment risk factor in a complex way. We develop a more global approach based on a Bayesian model that uses a latent genetic profile variable to capture all of the genetic variation in the entire targeted region and allows the environment effect to vary across different genetic profile categories. We also propose a resampling-based test derived from the developed Bayesian model for the detection of gene-environment interaction. Using data collected in the Environment and Genetics in Lung Cancer Etiology (EAGLE) study, we apply the Bayesian model to evaluate the joint effect of smoking intensity and genetic variants in the 15q25.1 region, which contains a cluster of nicotinic acetylcholine receptor genes and has been shown to be associated with both lung cancer and smoking behavior. We find evidence for gene-environment interaction (P-value = 0.016), with the smoking effect appearing to be stronger in subjects with a genetic profile associated with a higher lung cancer risk; the conventional test of gene-environment interaction based on the single-marker approach is far from significant.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
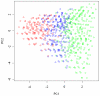

 for subjects in cluster 1, with the true value given by the horizontal line in green; (b). Boxplots of posterior medians of
for subjects in cluster 1, with the true value given by the horizontal line in green; (b). Boxplots of posterior medians of  for subjects in cluster 2, with the true value given by the horizontal line in blue. The posterior median of
for subjects in cluster 2, with the true value given by the horizontal line in blue. The posterior median of  for each subject under a given simulated dataset was shifted by a constant value selected so that the median value of the shifted estimates for subjects in cluster 1 was zero.
for each subject under a given simulated dataset was shifted by a constant value selected so that the median value of the shifted estimates for subjects in cluster 1 was zero.
 for subjects in cluster 1, with the true value given by the horizontal line in green; (b). Boxplots of posterior medians of
for subjects in cluster 1, with the true value given by the horizontal line in green; (b). Boxplots of posterior medians of  for subjects in cluster 2, with the true value given by the horizontal line in blue.
for subjects in cluster 2, with the true value given by the horizontal line in blue.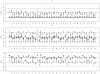
 for subjects in cluster 1, with the true value given by the horizontal line in green; (b). Boxplots of posterior medians of
for subjects in cluster 1, with the true value given by the horizontal line in green; (b). Boxplots of posterior medians of  for subjects in cluster 2, with the true value given by the horizontal line in blue; (c). Boxplots of posterior medians of
for subjects in cluster 2, with the true value given by the horizontal line in blue; (c). Boxplots of posterior medians of  for subjects in cluster 3, with the true value given by the horizontal line in red. The posterior median of
for subjects in cluster 3, with the true value given by the horizontal line in red. The posterior median of  for each subject under a given simulated dataset was shifted by a constant value selected so that the median value of the shifted estimates for subjects in cluster 1 was zero.
for each subject under a given simulated dataset was shifted by a constant value selected so that the median value of the shifted estimates for subjects in cluster 1 was zero.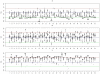
 for subjects in cluster 1, with the true value given by the horizontal line in green; (b). Boxplots of posterior medians of
for subjects in cluster 1, with the true value given by the horizontal line in green; (b). Boxplots of posterior medians of  for subjects in cluster 2, with the true value given by the horizontal line in blue; (c). Boxplots of posterior medians of
for subjects in cluster 2, with the true value given by the horizontal line in blue; (c). Boxplots of posterior medians of  for subjects in cluster 3, with the true value given by the horizontal line in red.
for subjects in cluster 3, with the true value given by the horizontal line in red.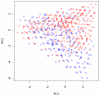
Similar articles
-
Commentary: gene-environment interactions and smoking-related cancers.Int J Epidemiol. 2010 Apr;39(2):577-9. doi: 10.1093/ije/dyp385. Epub 2010 Feb 1. Int J Epidemiol. 2010. PMID: 20123949 No abstract available.
-
Association between a 15q25 gene variant, smoking quantity and tobacco-related cancers among 17 000 individuals.Int J Epidemiol. 2010 Apr;39(2):563-77. doi: 10.1093/ije/dyp288. Epub 2009 Sep 23. Int J Epidemiol. 2010. PMID: 19776245 Free PMC article.
-
Gene by Environment Investigation of Incident Lung Cancer Risk in African-Americans.EBioMedicine. 2016 Jan 11;4:153-61. doi: 10.1016/j.ebiom.2016.01.002. eCollection 2016 Feb. EBioMedicine. 2016. PMID: 26981579 Free PMC article.
-
Contribution of Variants in CHRNA5/A3/B4 Gene Cluster on Chromosome 15 to Tobacco Smoking: From Genetic Association to Mechanism.Mol Neurobiol. 2016 Jan;53(1):472-484. doi: 10.1007/s12035-014-8997-x. Epub 2014 Dec 5. Mol Neurobiol. 2016. PMID: 25471942 Review.
-
Bayesian Approaches in Exploring Gene-environment and Gene-gene Interactions: A Comprehensive Review.Cancer Genomics Proteomics. 2023 Dec;20(6suppl):669-678. doi: 10.21873/cgp.20414. Cancer Genomics Proteomics. 2023. PMID: 38035701 Free PMC article. Review.
Cited by
-
Rat Mammary carcinoma susceptibility 3 (Mcs3) pleiotropy, socioenvironmental interaction, and comparative genomics with orthologous human 15q25.1-25.2.G3 (Bethesda). 2023 Jan 12;13(1):jkac288. doi: 10.1093/g3journal/jkac288. G3 (Bethesda). 2023. PMID: 36315068 Free PMC article.
-
Bayesian hierarchical models for high-dimensional mediation analysis with coordinated selection of correlated mediators.Stat Med. 2021 Nov 30;40(27):6038-6056. doi: 10.1002/sim.9168. Epub 2021 Aug 17. Stat Med. 2021. PMID: 34404112 Free PMC article.
-
Bivariate logistic Bayesian LASSO for detecting rare haplotype association with two correlated phenotypes.Genet Epidemiol. 2019 Dec;43(8):996-1017. doi: 10.1002/gepi.22258. Epub 2019 Sep 23. Genet Epidemiol. 2019. PMID: 31544985 Free PMC article.
-
Comparison of haplotype-based tests for detecting gene-environment interactions with rare variants.Brief Bioinform. 2020 May 21;21(3):851-862. doi: 10.1093/bib/bbz031. Brief Bioinform. 2020. PMID: 31329820 Free PMC article. Review.
-
The importance of gene-environment interactions in human obesity.Clin Sci (Lond). 2016 Sep 1;130(18):1571-97. doi: 10.1042/CS20160221. Clin Sci (Lond). 2016. PMID: 27503943 Free PMC article. Review.
References
-
- Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. A catalog of published genome-wide association studies. 2011. Available at: www.genome.gov/gwastudies. Accessed August, 2011.
-
- Lindstrom S, Schumacher F, Siddiq A, Travis RC, Campa D, et al. Characterizing associations and SNP-environment interactions for GWAS-identified prostate cancer risk markers-Results from BPC3. PLoS ONE. 2011;6:e17142. doi: 10.1371/journal.pone.0017142. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

