COMMD1 (copper metabolism MURR1 domain-containing protein 1) regulates Cullin RING ligases by preventing CAND1 (Cullin-associated Nedd8-dissociated protein 1) binding
- PMID: 21778237
- PMCID: PMC3173175
- DOI: 10.1074/jbc.M111.278408
COMMD1 (copper metabolism MURR1 domain-containing protein 1) regulates Cullin RING ligases by preventing CAND1 (Cullin-associated Nedd8-dissociated protein 1) binding
Abstract
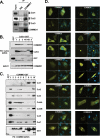
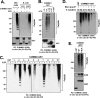
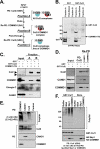
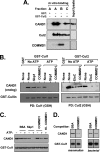
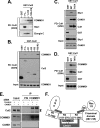
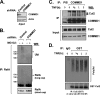
Cullin RING ligases (CRLs), the most prolific class of ubiquitin ligase enzymes, are multimeric complexes that regulate a wide range of cellular processes. CRL activity is regulated by CAND1 (Cullin-associated Nedd8-dissociated protein 1), an inhibitor that promotes the dissociation of substrate receptor components from the CRL. We demonstrate here that COMMD1 (copper metabolism MURR1 domain-containing 1), a factor previously found to promote ubiquitination of various substrates, regulates CRL activation by antagonizing CAND1 binding. We show that COMMD1 interacts with multiple Cullins, that the COMMD1-Cul2 complex cannot bind CAND1, and that, conversely, COMMD1 can actively displace CAND1 from CRLs. These findings highlight a novel mechanism of CRL activation and suggest that CRL regulation may underlie the pleiotropic activities of COMMD1.
Figures
Similar articles
-
Association of SAP130/SF3b-3 with Cullin-RING ubiquitin ligase complexes and its regulation by the COP9 signalosome.BMC Biochem. 2008 Jan 3;9:1. doi: 10.1186/1471-2091-9-1. BMC Biochem. 2008. PMID: 18173839 Free PMC article.
-
Drosophila Cand1 regulates Cullin3-dependent E3 ligases by affecting the neddylation of Cullin3 and by controlling the stability of Cullin3 and adaptor protein.Dev Biol. 2010 Oct 15;346(2):247-57. doi: 10.1016/j.ydbio.2010.07.031. Epub 2010 Aug 4. Dev Biol. 2010. PMID: 20691177
-
Regulation of cullin RING E3 ubiquitin ligases by CAND1 in vivo.PLoS One. 2011 Jan 13;6(1):e16071. doi: 10.1371/journal.pone.0016071. PLoS One. 2011. PMID: 21249194 Free PMC article.
-
Assembly and Regulation of CRL Ubiquitin Ligases.Adv Exp Med Biol. 2020;1217:33-46. doi: 10.1007/978-981-15-1025-0_3. Adv Exp Med Biol. 2020. PMID: 31898220 Review.
-
Deregulation of the COP9 signalosome-cullin-RING ubiquitin-ligase pathway: mechanisms and roles in urological cancers.Int J Biochem Cell Biol. 2013 Jul;45(7):1327-37. doi: 10.1016/j.biocel.2013.03.023. Epub 2013 Apr 10. Int J Biochem Cell Biol. 2013. PMID: 23583660 Review.
Cited by
-
Knockdown of cullin 3 inhibits progressive phenotypes and increases chemosensitivity in cholangiocarcinoma cells.Mol Med Rep. 2024 Nov;30(5):198. doi: 10.3892/mmr.2024.13322. Epub 2024 Sep 6. Mol Med Rep. 2024. PMID: 39239747 Free PMC article.
-
Systems-wide Studies Uncover Commander, a Multiprotein Complex Essential to Human Development.Cell Syst. 2017 May 24;4(5):483-494. doi: 10.1016/j.cels.2017.04.006. Cell Syst. 2017. PMID: 28544880 Free PMC article. Review.
-
CCDC22 deficiency in humans blunts activation of proinflammatory NF-κB signaling.J Clin Invest. 2013 May;123(5):2244-56. doi: 10.1172/JCI66466. Epub 2013 Apr 8. J Clin Invest. 2013. PMID: 23563313 Free PMC article.
-
Commander Complex-A Multifaceted Operator in Intracellular Signaling and Cargo.Cells. 2021 Dec 7;10(12):3447. doi: 10.3390/cells10123447. Cells. 2021. PMID: 34943955 Free PMC article. Review.
-
The Copper Metabolism MURR1 domain protein 1 (COMMD1) modulates the aggregation of misfolded protein species in a client-specific manner.PLoS One. 2014 Apr 1;9(4):e92408. doi: 10.1371/journal.pone.0092408. eCollection 2014. PLoS One. 2014. PMID: 24691167 Free PMC article.
References
-
- Weissman A. M. (2001) Nat. Rev. Mol. Cell Biol. 2, 169–178 - PubMed
-
- Wilkinson C. R. (2002) Trends Cell Biol. 12, 545–546 - PubMed
-
- Joazeiro C. A., Weissman A. M. (2000) Cell 102, 549–552 - PubMed
-
- Patterson C. (2002) Sci. STKE 2002, pe4. - PubMed
-
- Coscoy L., Ganem D. (2003) Trends Cell Biol. 13, 7–12 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

