Dali server: conservation mapping in 3D
- PMID: 20457744
- PMCID: PMC2896194
- DOI: 10.1093/nar/gkq366
Dali server: conservation mapping in 3D
Abstract
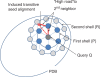
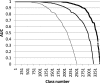
Our web site (http://ekhidna.biocenter.helsinki.fi/dali_server) runs the Dali program for protein structure comparison. The web site consists of three parts: (i) the Dali server compares newly solved structures against structures in the Protein Data Bank (PDB), (ii) the Dali database allows browsing precomputed structural neighbourhoods and (iii) the pairwise comparison generates suboptimal alignments for a pair of structures. Each part has its own query form and a common format for the results page. The inputs are either PDB identifiers or novel structures uploaded by the user. The results pages are hyperlinked to aid interactive analysis. The web interface is simple and easy to use. The key purpose of interactive analysis is to check whether conserved residues line up in multiple structural alignments and how conserved residues and ligands cluster together in multiple structure superimpositions. In favourable cases, protein structure comparison can lead to evolutionary discoveries not detected by sequence analysis.
Figures

Similar articles
-
Dali server update.Nucleic Acids Res. 2016 Jul 8;44(W1):W351-5. doi: 10.1093/nar/gkw357. Epub 2016 Apr 29. Nucleic Acids Res. 2016. PMID: 27131377 Free PMC article.
-
Using Dali for Protein Structure Comparison.Methods Mol Biol. 2020;2112:29-42. doi: 10.1007/978-1-0716-0270-6_3. Methods Mol Biol. 2020. PMID: 32006276
-
Searching protein structure databases with DaliLite v.3.Bioinformatics. 2008 Dec 1;24(23):2780-1. doi: 10.1093/bioinformatics/btn507. Epub 2008 Sep 25. Bioinformatics. 2008. PMID: 18818215 Free PMC article.
-
QSalignWeb: A Server to Predict and Analyze Protein Quaternary Structure.Front Mol Biosci. 2022 Jan 5;8:787510. doi: 10.3389/fmolb.2021.787510. eCollection 2021. Front Mol Biosci. 2022. PMID: 35071324 Free PMC article. Review.
-
Comparison of proteins based on segments structural similarity.Acta Biochim Pol. 2004;51(1):161-72. Acta Biochim Pol. 2004. PMID: 15094837 Review.
Cited by
-
Structural Insights into the Effector - Immunity System Tae4/Tai4 from Salmonella typhimurium.PLoS One. 2013 Jun 27;8(6):e67362. doi: 10.1371/journal.pone.0067362. Print 2013. PLoS One. 2013. PMID: 23826277 Free PMC article.
-
The AEROPATH project targeting Pseudomonas aeruginosa: crystallographic studies for assessment of potential targets in early-stage drug discovery.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Jan 1;69(Pt 1):25-34. doi: 10.1107/S1744309112044739. Epub 2012 Dec 25. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23295481 Free PMC article.
-
Domain organization of DNase from Thioalkalivibrio sp. provides insights into retention of activity in high salt environments.Front Microbiol. 2015 Jul 1;6:661. doi: 10.3389/fmicb.2015.00661. eCollection 2015. Front Microbiol. 2015. PMID: 26191053 Free PMC article.
-
Structure of Vibrio cholerae ribosome hibernation promoting factor.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013 Mar 1;69(Pt 3):228-36. doi: 10.1107/S1744309113000961. Epub 2013 Feb 22. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2013. PMID: 23519794 Free PMC article.
-
Structural insights into the functional role of the Hcn sub-domain of the receptor-binding domain of the botulinum neurotoxin mosaic serotype C/D.Biochimie. 2013 Jul;95(7):1379-85. doi: 10.1016/j.biochi.2013.03.006. Epub 2013 Mar 21. Biochimie. 2013. PMID: 23523511 Free PMC article.
References
-
- Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:381–389. - PubMed
-
- Kawabata T, Nishikawa K. Protein tertiary structure comparison using the Markov transition model of evolution. Proteins. 2000;41:108–122. - PubMed
-
- Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst. 2004;D60:2256–2268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

