X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface
- PMID: 19846792
- PMCID: PMC2765457
- DOI: 10.1073/pnas.0907990106
X-ray structure of Pur-alpha reveals a Whirly-like fold and an unusual nucleic-acid binding surface
Abstract
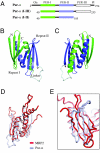
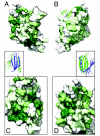
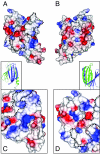
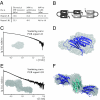
The PUR protein family is a distinct and highly conserved class that is characterized by its sequence-specific RNA- and DNA-binding. Its best-studied family member, Pur-alpha, acts as a transcriptional regulator, as host factor for viral replication, and as cofactor for mRNP localization in dendrites. Pur-alpha-deficient mice show severe neurologic defects and die after birth. Nucleic-acid binding by Pur-alpha is mediated by its central core region, for which no structural information is available. We determined the x-ray structure of residues 40 to 185 from Drosophila melanogaster Pur-alpha, which constitutes a major part of the core region. We found that this region contains two almost identical structural motifs, termed "PUR repeats," which interact with each other to form a PUR domain. DNA- and RNA-binding studies confirmed that PUR domains are indeed functional nucleic-acid binding domains. Database analysis show that PUR domains share a fold with the Whirly class of nucleic-acid binding proteins. Structural analysis combined with mutational studies suggest that a PUR domain binds nucleic acids through two independent surface regions involving concave beta-sheets. Structure-based sequence alignment revealed that the core region harbors a third PUR repeat at its C terminus. Subsequent characterization by small-angle x-ray scattering (SAXS) and size-exclusion chromatography indicated that PUR repeat III mediates dimerization of Pur-alpha. Surface envelopes calculated from SAXS data show that the Pur-alpha dimer consisting of repeats I to III is arranged in a Z-like shape. This unexpected domain organization of the entire core domain of Pur-alpha has direct implications for ssDNA/ssRNA and dsDNA binding.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Structural basis of nucleic-acid recognition and double-strand unwinding by the essential neuronal protein Pur-alpha.Elife. 2016 Jan 8;5:e11297. doi: 10.7554/eLife.11297. Elife. 2016. PMID: 26744780 Free PMC article.
-
Drosophila Pur-α binds to trinucleotide-repeat containing cellular RNAs and translocates to the early oocyte.RNA Biol. 2012 May;9(5):633-43. doi: 10.4161/rna.19760. Epub 2012 May 1. RNA Biol. 2012. PMID: 22614836
-
PURA, the gene encoding Pur-alpha, member of an ancient nucleic acid-binding protein family with mammalian neurological functions.Gene. 2018 Feb 15;643:133-143. doi: 10.1016/j.gene.2017.12.004. Epub 2017 Dec 6. Gene. 2018. PMID: 29221753 Free PMC article. Review.
-
Nucleoplasmin-like domain of FKBP39 from Drosophila melanogaster forms a tetramer with partly disordered tentacle-like C-terminal segments.Sci Rep. 2017 Jan 11;7:40405. doi: 10.1038/srep40405. Sci Rep. 2017. PMID: 28074868 Free PMC article.
-
Integrated structural biology to unravel molecular mechanisms of protein-RNA recognition.Methods. 2017 Apr 15;118-119:119-136. doi: 10.1016/j.ymeth.2017.03.015. Epub 2017 Mar 16. Methods. 2017. PMID: 28315749 Review.
Cited by
-
Small-angle scattering for structural biology--expanding the frontier while avoiding the pitfalls.Protein Sci. 2010 Apr;19(4):642-57. doi: 10.1002/pro.351. Protein Sci. 2010. PMID: 20120026 Free PMC article. Review.
-
Acquisition of an Archaea-like ribonuclease H domain by plant L1 retrotransposons supports modular evolution.Proc Natl Acad Sci U S A. 2013 Dec 10;110(50):20140-5. doi: 10.1073/pnas.1310958110. Epub 2013 Nov 25. Proc Natl Acad Sci U S A. 2013. PMID: 24277848 Free PMC article.
-
Posttranscriptional self-regulation by the Lyme disease bacterium's BpuR DNA/RNA-binding protein.J Bacteriol. 2013 Nov;195(21):4915-23. doi: 10.1128/JB.00819-13. Epub 2013 Aug 23. J Bacteriol. 2013. PMID: 23974034 Free PMC article.
-
Post-translational modifications regulate assembly of early spindle orientation complex in yeast.J Biol Chem. 2012 May 11;287(20):16238-45. doi: 10.1074/jbc.M112.347872. Epub 2012 Mar 29. J Biol Chem. 2012. PMID: 22461628 Free PMC article.
-
Structural and functional analysis of single-nucleotide polymorphic variants of purine-rich element-binding protein B.J Cell Biochem. 2019 Apr;120(4):5835-5851. doi: 10.1002/jcb.27869. Epub 2018 Nov 1. J Cell Biochem. 2019. PMID: 30387171 Free PMC article.
References
-
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: Isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. - PubMed
-
- Ohashi S, et al. Identification of mRNA/protein (mRNP) complexes containing Pur-alpha, mStaufen, fragile X protein, and myosin Va and their association with rough endoplasmic reticulum equipped with a kinesin motor. J Biol Chem. 2002;277:37804–37810. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

