Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard
- PMID: 19465773
- PMCID: PMC2685735
- DOI: 10.1107/S0907444909012098
Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard
Abstract
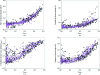
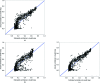
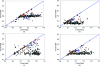
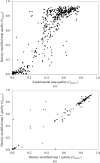
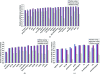
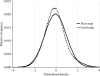
Estimates of the quality of experimental maps are important in many stages of structure determination of macromolecules. Map quality is defined here as the correlation between a map and the corresponding map obtained using phases from the final refined model. Here, ten different measures of experimental map quality were examined using a set of 1359 maps calculated by re-analysis of 246 solved MAD, SAD and MIR data sets. A simple Bayesian approach to estimation of map quality from one or more measures is presented. It was found that a Bayesian estimator based on the skewness of the density values in an electron-density map is the most accurate of the ten individual Bayesian estimators of map quality examined, with a correlation between estimated and actual map quality of 0.90. A combination of the skewness of electron density with the local correlation of r.m.s. density gives a further improvement in estimating map quality, with an overall correlation coefficient of 0.92. The PHENIX AutoSol wizard carries out automated structure solution based on any combination of SAD, MAD, SIR or MIR data sets. The wizard is based on tools from the PHENIX package and uses the Bayesian estimates of map quality described here to choose the highest quality solutions after experimental phasing.
Figures


Similar articles
-
Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard.Acta Crystallogr D Biol Crystallogr. 2008 Jan;64(Pt 1):61-9. doi: 10.1107/S090744490705024X. Epub 2007 Dec 5. Acta Crystallogr D Biol Crystallogr. 2008. PMID: 18094468 Free PMC article.
-
Can I solve my structure by SAD phasing? Planning an experiment, scaling data and evaluating the useful anomalous correlation and anomalous signal.Acta Crystallogr D Struct Biol. 2016 Mar;72(Pt 3):359-74. doi: 10.1107/S2059798315019403. Epub 2016 Mar 1. Acta Crystallogr D Struct Biol. 2016. PMID: 26960123 Free PMC article.
-
RNA Structure Refinement Using the ERRASER-Phenix Pipeline.Methods Mol Biol. 2016;1320:269-82. doi: 10.1007/978-1-4939-2763-0_17. Methods Mol Biol. 2016. PMID: 26227049 Free PMC article.
-
Experimental Phasing: Substructure Solution and Density Modification as Implemented in SHELX.Methods Mol Biol. 2017;1607:357-376. doi: 10.1007/978-1-4939-7000-1_15. Methods Mol Biol. 2017. PMID: 28573581 Review.
-
You are lost without a map: Navigating the sea of protein structures.Biochim Biophys Acta. 2015 Apr;1854(4):258-68. doi: 10.1016/j.bbapap.2014.12.021. Epub 2014 Dec 29. Biochim Biophys Acta. 2015. PMID: 25554228 Free PMC article. Review.
Cited by
-
Crystal structure of SsfS6, the putative C-glycosyltransferase involved in SF2575 biosynthesis.Proteins. 2013 Jul;81(7):1277-82. doi: 10.1002/prot.24289. Epub 2013 Apr 20. Proteins. 2013. PMID: 23526584 Free PMC article.
-
Determining crystal structures through crowdsourcing and coursework.Nat Commun. 2016 Sep 16;7:12549. doi: 10.1038/ncomms12549. Nat Commun. 2016. PMID: 27633552 Free PMC article.
-
Active site and laminarin binding in glycoside hydrolase family 55.J Biol Chem. 2015 May 8;290(19):11819-32. doi: 10.1074/jbc.M114.623579. Epub 2015 Mar 9. J Biol Chem. 2015. PMID: 25752603 Free PMC article.
-
Structure, dynamics, and stability of the smallest and most complex 71 protein knot.J Biol Chem. 2024 Jan;300(1):105553. doi: 10.1016/j.jbc.2023.105553. Epub 2023 Dec 10. J Biol Chem. 2024. PMID: 38072060 Free PMC article.
-
Structural basis of flagellar motility regulation by the MogR repressor and the GmaR antirepressor in Listeria monocytogenes.Nucleic Acids Res. 2022 Oct 28;50(19):11315-11330. doi: 10.1093/nar/gkac815. Nucleic Acids Res. 2022. PMID: 36283692 Free PMC article.
References
-
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. - PubMed
-
- Alphey, M. S., Leonard, G. A., Gourley, D. G., Tetaud, E., Fairlamb, A. H. & Hunter, W. N. (1999). J. Biol. Chem.274, 25613–25622. - PubMed
-
- Baker, D., Bystroff, C., Fletterick, R. J. & Agard, D. A. (1993). Acta Cryst. D49, 429–439. - PubMed
-
- Bernstein, F. C., Koetzle, T. F., Williams, G. J. B., Meyer, E. F. Jr, Brice, M. D., Rodgers, J. R., Kennard, O., Shimanouchi, T. & Tasumi, M. (1977). J. Mol. Biol.112, 535–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

