COMMD1 expression is controlled by critical residues that determine XIAP binding
- PMID: 18795889
- PMCID: PMC2606926
- DOI: 10.1042/BJ20080854
COMMD1 expression is controlled by critical residues that determine XIAP binding
Abstract
COMMD {COMM [copper metabolism Murr1 (mouse U2af1-rs1 region 1)] domain-containing} proteins participate in several cellular processes, ranging from NF-kappaB (nuclear factor kappaB) regulation, copper homoeostasis, sodium transport and adaptation to hypoxia. The best-studied member of this family is COMMD1, but relatively little is known about its regulation, except that XIAP [X-linked IAP (inhibitor of apoptosis)] functions as its ubiquitin ligase. In the present study, we identified that the COMM domain of COMMD1 is required for its interaction with XIAP, and other COMMD proteins can similarly interact with IAPs. Two conserved leucine repeats within the COMM domain were found to be critically required for XIAP binding. A COMMD1 mutant which was unable to bind to XIAP demonstrated a complete loss of basal ubiquitination and great stabilization of the protein. Underscoring the importance of IAP-mediated ubiquitination, we found that long-term expression of wild-type COMMD1 results in nearly physiological protein levels as a result of increased ubiquitination, but this regulatory event is circumvented when a mutant form that cannot bind XIAP is expressed. In summary, our findings indicate that COMMD1 expression is controlled primarily by protein ubiquitination, and its interaction with IAP proteins plays an essential role.
Figures
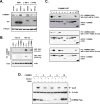
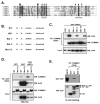
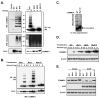

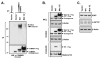
Similar articles
-
Characterization of COMMD protein-protein interactions in NF-kappaB signalling.Biochem J. 2006 Aug 15;398(1):63-71. doi: 10.1042/BJ20051664. Biochem J. 2006. PMID: 16573520 Free PMC article.
-
Characterization and copper binding properties of human COMMD1 (MURR1).Biochemistry. 2007 Mar 20;46(11):3116-28. doi: 10.1021/bi0620656. Epub 2007 Feb 20. Biochemistry. 2007. PMID: 17309234
-
p300-mediated acetylation of COMMD1 regulates its stability, and the ubiquitylation and nucleolar translocation of the RelA NF-κB subunit.J Cell Sci. 2014 Sep 1;127(Pt 17):3659-65. doi: 10.1242/jcs.149328. Epub 2014 Jul 29. J Cell Sci. 2014. PMID: 25074812 Free PMC article.
-
COMMD proteins and the control of the NF kappa B pathway.Cell Cycle. 2007 Mar 15;6(6):672-6. doi: 10.4161/cc.6.6.3989. Epub 2007 Mar 7. Cell Cycle. 2007. PMID: 17361106 Free PMC article. Review.
-
XIAP as a ubiquitin ligase in cellular signaling.Cell Death Differ. 2010 Jan;17(1):54-60. doi: 10.1038/cdd.2009.81. Cell Death Differ. 2010. PMID: 19590513 Free PMC article. Review.
Cited by
-
Copper homeostasis and the ubiquitin proteasome system.Metallomics. 2023 Mar 6;15(3):mfad010. doi: 10.1093/mtomcs/mfad010. Metallomics. 2023. PMID: 36822629 Free PMC article. Review.
-
Posttranslational regulation of copper transporters.J Biol Inorg Chem. 2010 Jan;15(1):37-46. doi: 10.1007/s00775-009-0592-7. Epub 2009 Oct 8. J Biol Inorg Chem. 2010. PMID: 19813030 Review.
-
The Inhibitor of Apoptosis (IAPs) in Adaptive Response to Cellular Stress.Cells. 2012 Oct 10;1(4):711-37. doi: 10.3390/cells1040711. Cells. 2012. PMID: 24710527 Free PMC article.
-
COMMD1 forms oligomeric complexes targeted to the endocytic membranes via specific interactions with phosphatidylinositol 4,5-bisphosphate.J Biol Chem. 2009 Jan 2;284(1):696-707. doi: 10.1074/jbc.M804766200. Epub 2008 Oct 21. J Biol Chem. 2009. PMID: 18940794 Free PMC article.
-
Silver Ions as a Tool for Understanding Different Aspects of Copper Metabolism.Nutrients. 2019 Jun 17;11(6):1364. doi: 10.3390/nu11061364. Nutrients. 2019. PMID: 31213024 Free PMC article. Review.
References
-
- Burstein E, Hoberg JE, Wilkinson AS, Rumble JM, Csomos RA, Komarck CM, Maine GN, Wilkinson JC, Mayo MW, Duckett CS. COMMD proteins: A novel family of structural and functional homologs of MURR1. J Biol Chem. 2005;280:22222–22232. - PubMed
-
- Narindrasorasak S, Kulkarni P, Deschamps P, She YM, Sarkar B. Characterization and Copper Binding Properties of Human COMMD1 (MURR1) Biochemistry. 2007;46:3116–3128. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

