Identification of cross-linked peptides from large sequence databases
- PMID: 18327264
- PMCID: PMC2719781
- DOI: 10.1038/nmeth.1192
Identification of cross-linked peptides from large sequence databases
Erratum in
- Nat Methods. 2008 Aug;5(8):748
Abstract
We describe a method to identify cross-linked peptides from complex samples and large protein sequence databases by combining isotopically tagged cross-linkers, chromatographic enrichment, targeted proteomics and a new search engine called xQuest. This software reduces the search space by an upstream candidate-peptide search before the recombination step. We showed that xQuest can identify cross-linked peptides from a total Escherichia coli lysate with an unrestricted database search.
Figures
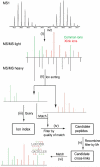
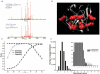
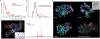
Similar articles
-
Identification of cross-linked peptides from complex samples.Nat Methods. 2012 Sep;9(9):904-6. doi: 10.1038/nmeth.2099. Epub 2012 Jul 8. Nat Methods. 2012. PMID: 22772728
-
ECL: an exhaustive search tool for the identification of cross-linked peptides using whole database.BMC Bioinformatics. 2016 May 20;17(1):217. doi: 10.1186/s12859-016-1073-y. BMC Bioinformatics. 2016. PMID: 27206479 Free PMC article.
-
False discovery rate estimation for cross-linked peptides identified by mass spectrometry.Nat Methods. 2012 Sep;9(9):901-3. doi: 10.1038/nmeth.2103. Epub 2012 Jul 8. Nat Methods. 2012. PMID: 22772729
-
Cross-linked peptide identification: A computational forest of algorithms.Mass Spectrom Rev. 2018 Nov;37(6):738-749. doi: 10.1002/mas.21559. Epub 2018 Mar 12. Mass Spectrom Rev. 2018. PMID: 29529716 Review.
-
Cross-Linking Mass Spectrometry: An Emerging Technology for Interactomics and Structural Biology.Anal Chem. 2018 Jan 2;90(1):144-165. doi: 10.1021/acs.analchem.7b04431. Epub 2017 Nov 21. Anal Chem. 2018. PMID: 29160693 Free PMC article. Review.
Cited by
-
Ensemble-based design of tau to inhibit aggregation while preserving biological activity.Res Sq [Preprint]. 2024 Jan 16:rs.3.rs-3796916. doi: 10.21203/rs.3.rs-3796916/v1. Res Sq. 2024. PMID: 38313287 Free PMC article. Preprint.
-
Quantitative cross-linking/mass spectrometry using isotope-labelled cross-linkers.J Proteomics. 2013 Aug 2;88:120-8. doi: 10.1016/j.jprot.2013.03.005. Epub 2013 Mar 26. J Proteomics. 2013. PMID: 23541715 Free PMC article.
-
In vivo protein complex topologies: sights through a cross-linking lens.Proteomics. 2012 May;12(10):1565-75. doi: 10.1002/pmic.201100516. Proteomics. 2012. PMID: 22610688 Free PMC article. Review.
-
Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography.Mol Cell Proteomics. 2012 Mar;11(3):M111.014126. doi: 10.1074/mcp.M111.014126. Epub 2012 Jan 27. Mol Cell Proteomics. 2012. PMID: 22286754 Free PMC article.
-
Proteomics-based methods for discovery, quantification, and validation of protein-protein interactions.Anal Chem. 2013 Jan 15;85(2):749-68. doi: 10.1021/ac3033257. Epub 2012 Dec 12. Anal Chem. 2013. PMID: 23157382 Free PMC article. Review. No abstract available.
References
-
- Sinz A. Chemical cross-linking and mass spectrometry for mapping three-dimensional structures of proteins and protein complexes. J Mass Spectrom. 2003;38:1225–1237. - PubMed
-
- Seebacher J, et al. Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. Journal of proteome research. 2006;5:2270–2282. - PubMed
-
- Ihling C, et al. Isotope-Labeled Cross-Linkers and Fourier Transform Ion Cyclotron Resonance Mass Spectrometry for Structural Analysis of a Protein/Peptide Complex. J Am Soc Mass Spectrom. 2006 - PubMed
-
- Muller DR, et al. Isotope-tagged cross-linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal Chem. 2001;73:1927–1934. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

