Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice
- PMID: 17371845
- PMCID: PMC1900009
- DOI: 10.1128/MCB.01932-06
Increased activity of hypoxia-inducible factor 1 is associated with early embryonic lethality in Commd1 null mice
Abstract
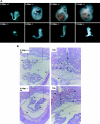
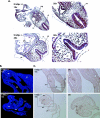
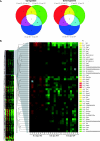
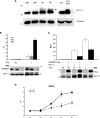
COMMD1 (previously known as MURR1) belongs to a novel family of proteins termed the copper metabolism gene MURR1 domain (COMMD) family. The 10 COMMD family members are well conserved between vertebrates, but the functions of most of the COMMD proteins are unknown. We recently established that COMMD1 is associated with the hepatic copper overload disorder copper toxicosis in Bedlington terriers. Recent in vitro studies indicate that COMMD1 has multiple functions, including sodium transport and NF-kappaB signaling. To elucidate the function of Commd1 in vivo, we generated homozygous Commd1 null (Commd1(-/-)) mice. Commd1(-/-) embryos died in utero between 9.5 and 10.5 days postcoitum (dpc), their development was generally retarded, and placenta vascularization was absent. Microarray analysis identified transcriptional upregulation of hypoxia-inducible factor 1 (HIF-1) target genes in 9.5-dpc Commd1(-/-) embryos compared to normal embryos, a feature that was associated with increased Hif-1alpha stability. Consistent with these observations, COMMD1 physically associates with HIF-1alpha and inhibits HIF-1alpha stability and HIF-1 transactivation in vitro. Thus, this study identifies COMMD1 as a novel regulator of HIF-1 activity and shows that Commd1 deficiency in mice leads to embryonic lethality associated with dysregulated placenta vascularization.
Figures


Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Antioxidants for female subfertility.Cochrane Database Syst Rev. 2020 Aug 27;8(8):CD007807. doi: 10.1002/14651858.CD007807.pub4. Cochrane Database Syst Rev. 2020. PMID: 32851663 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Genedrive kit for detecting single nucleotide polymorphism m.1555A>G in neonates and their mothers: a systematic review and cost-effectiveness analysis.Health Technol Assess. 2024 Oct;28(75):1-75. doi: 10.3310/TGAC4201. Health Technol Assess. 2024. PMID: 39487741 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
Cited by
-
Expression and localization of COMMD1 proteins in human placentas from women with preeclampsia.Yonsei Med J. 2013 Mar 1;54(2):494-9. doi: 10.3349/ymj.2013.54.2.494. Yonsei Med J. 2013. PMID: 23364987 Free PMC article.
-
Liver-specific Commd1 knockout mice are susceptible to hepatic copper accumulation.PLoS One. 2011;6(12):e29183. doi: 10.1371/journal.pone.0029183. Epub 2011 Dec 22. PLoS One. 2011. PMID: 22216203 Free PMC article.
-
COMMD1 (copper metabolism MURR1 domain-containing protein 1) regulates Cullin RING ligases by preventing CAND1 (Cullin-associated Nedd8-dissociated protein 1) binding.J Biol Chem. 2011 Sep 16;286(37):32355-65. doi: 10.1074/jbc.M111.278408. Epub 2011 Jul 21. J Biol Chem. 2011. PMID: 21778237 Free PMC article.
-
Systems-wide Studies Uncover Commander, a Multiprotein Complex Essential to Human Development.Cell Syst. 2017 May 24;4(5):483-494. doi: 10.1016/j.cels.2017.04.006. Cell Syst. 2017. PMID: 28544880 Free PMC article. Review.
-
Hypoxia and Placental Development.Birth Defects Res. 2017 Oct 16;109(17):1309-1329. doi: 10.1002/bdr2.1135. Birth Defects Res. 2017. PMID: 29105383 Free PMC article. Review.
References
-
- Aprelikova, O., G. V. Chandramouli, M. Wood, J. R. Vasselli, J. Riss, J. K. Maranchie, W. M. Linehan, and J. C. Barrett. 2004. Regulation of HIF prolyl hydroxylases by hypoxia-inducible factors. J. Cell. Biochem. 92:491-501. - PubMed
-
- Biasio, W., T. Chang, C. J. McIntosh, and F. J. McDonald. 2004. Identification of Murr1 as a regulator of the human delta epithelial sodium channel. J. Biol. Chem. 279:5429-5434. - PubMed
-
- Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2:243-247. - PubMed
-
- Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
