Global rigid body modeling of macromolecular complexes against small-angle scattering data
- PMID: 15923225
- PMCID: PMC1366608
- DOI: 10.1529/biophysj.105.064154
Global rigid body modeling of macromolecular complexes against small-angle scattering data
Abstract
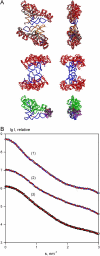
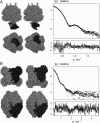
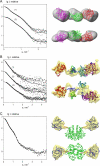
New methods to automatically build models of macromolecular complexes from high-resolution structures or homology models of their subunits or domains against x-ray or neutron small-angle scattering data are presented. Depending on the complexity of the object, different approaches are employed for the global search of the optimum configuration of subunits fitting the experimental data. An exhaustive grid search is used for hetero- and homodimeric particles and for symmetric oligomers formed by identical subunits. For the assemblies or multidomain proteins containing more then one subunit/domain per asymmetric unit, heuristic algorithms based on simulated annealing are used. Fast computational algorithms based on spherical harmonics representation of scattering amplitudes are employed. The methods allow one to construct interconnected models without steric clashes, to account for the particle symmetry and to incorporate information from other methods, on distances between specific residues or nucleotides. For multidomain proteins, addition of missing linkers between the domains is possible. Simultaneous fitting of multiple scattering patterns from subcomplexes or deletion mutants is incorporated. The efficiency of the methods is illustrated by their application to complexes of different types in several simulated and practical examples. Limitations and possible ambiguity of rigid body modeling are discussed and simplified docking criteria are provided to rank multiple models. The methods described are implemented in publicly available computer programs running on major hardware platforms.
Figures
Similar articles
-
Joint use of small-angle X-ray and neutron scattering to study biological macromolecules in solution.Eur Biophys J. 2006 Sep;35(7):567-76. doi: 10.1007/s00249-006-0063-9. Epub 2006 Apr 25. Eur Biophys J. 2006. PMID: 16636827
-
Addition of missing loops and domains to protein models by x-ray solution scattering.Biophys J. 2002 Dec;83(6):3113-25. doi: 10.1016/S0006-3495(02)75315-0. Biophys J. 2002. PMID: 12496082 Free PMC article.
-
Small-angle scattering and neutron contrast variation for studying bio-molecular complexes.Methods Mol Biol. 2009;544:307-23. doi: 10.1007/978-1-59745-483-4_20. Methods Mol Biol. 2009. PMID: 19488708
-
Structural studies of RNA-protein complexes: A hybrid approach involving hydrodynamics, scattering, and computational methods.Methods. 2017 Apr 15;118-119:146-162. doi: 10.1016/j.ymeth.2016.12.002. Epub 2016 Dec 8. Methods. 2017. PMID: 27939506 Review.
-
Analysis of X-ray and neutron scattering from biomacromolecular solutions.Curr Opin Struct Biol. 2007 Oct;17(5):562-71. doi: 10.1016/j.sbi.2007.06.009. Epub 2007 Aug 21. Curr Opin Struct Biol. 2007. PMID: 17714935 Review.
Cited by
-
Peroxisomal proteostasis involves a Lon family protein that functions as protease and chaperone.J Biol Chem. 2012 Aug 10;287(33):27380-95. doi: 10.1074/jbc.M112.381566. Epub 2012 Jun 25. J Biol Chem. 2012. PMID: 22733816 Free PMC article.
-
Molecular determinants of MED1 interaction with the DNA bound VDR-RXR heterodimer.Nucleic Acids Res. 2020 Nov 4;48(19):11199-11213. doi: 10.1093/nar/gkaa775. Nucleic Acids Res. 2020. PMID: 32990725 Free PMC article.
-
Hsp90 regulates the dynamics of its cochaperone Sti1 and the transfer of Hsp70 between modules.Nat Commun. 2015 Apr 8;6:6655. doi: 10.1038/ncomms7655. Nat Commun. 2015. PMID: 25851214 Free PMC article.
-
Recognition of viral RNA stem-loops by the tandem double-stranded RNA binding domains of PKR.RNA. 2013 Mar;19(3):333-44. doi: 10.1261/rna.035931.112. Epub 2013 Jan 17. RNA. 2013. PMID: 23329698 Free PMC article.
-
The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors.RNA. 2012 Nov;18(11):2056-72. doi: 10.1261/rna.034181.112. Epub 2012 Sep 20. RNA. 2012. PMID: 22996915 Free PMC article.
References
-
- Gerstein, M., A. Edwards, C. H. Arrowsmith, and G. T. Montelione. 2003. Structural genomics: current progress. Science. 299:1663. - PubMed
-
- Sali, A., R. Glaeser, T. Earnest, and W. Baumeister. 2003. From words to literature in structural proteomics. Nature. 422:216–225. - PubMed
-
- Aloy, P., B. Bottcher, H. Ceulemans, C. Leutwein, C. Mellwig, S. Fischer, A. C. Gavin, P. Bork, G. Superti-Furga, L. Serrano, and R. B. Russell. 2004. Structure-based assembly of protein complexes in yeast. Science. 303:2026–2029. - PubMed
-
- Feigin, L. A., and D. I. Svergun. 1987. Structure analysis by small-angle x-ray and neutron scattering. Plenum Press, New York.
-
- Svergun, D. I., and M. H. J. Koch. 2003. Small angle scattering studies of biological macromolecules in solution. Rep. Prog. Phys. 66:1735–1782.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

