σ28-dependent small RNA regulation of flagella biosynthesis
- PMID: 37843988
- PMCID: PMC10578931
- DOI: 10.7554/eLife.87151
σ28-dependent small RNA regulation of flagella biosynthesis
Abstract
Flagella are important for bacterial motility as well as for pathogenesis. Synthesis of these structures is energy intensive and, while extensive transcriptional regulation has been described, little is known about the posttranscriptional regulation. Small RNAs (sRNAs) are widespread posttranscriptional regulators, most base pairing with mRNAs to affect their stability and/or translation. Here, we describe four UTR-derived sRNAs (UhpU, MotR, FliX and FlgO) whose expression is controlled by the flagella sigma factor σ28 (fliA) in Escherichia coli. Interestingly, the four sRNAs have varied effects on flagellin protein levels, flagella number and cell motility. UhpU, corresponding to the 3´ UTR of a metabolic gene, likely has hundreds of targets including a transcriptional regulator at the top flagella regulatory cascade connecting metabolism and flagella synthesis. Unlike most sRNAs, MotR and FliX base pair within the coding sequences of target mRNAs and act on ribosomal protein mRNAs connecting ribosome production and flagella synthesis. The study shows how sRNA-mediated regulation can overlay a complex network enabling nuanced control of flagella synthesis.
Keywords: E. coli; Hfq; RIL-seq; S10; flagella; fliA; genetics; genomics; infectious disease; microbiology; post-transcriptional regulation.
Conflict of interest statement
SM, AZ, MJ, JM, AS, HZ, GS No competing interests declared
Figures

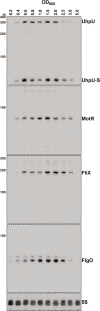










Update of
- doi: 10.1101/2021.08.05.455139
- doi: 10.7554/eLife.87151.1
- doi: 10.7554/eLife.87151.2
Similar articles
-
Dynamic interactions between the RNA chaperone Hfq, small regulatory RNAs, and mRNAs in live bacterial cells.Elife. 2021 Feb 22;10:e64207. doi: 10.7554/eLife.64207. Elife. 2021. PMID: 33616037 Free PMC article.
-
Competition among Hfq-binding small RNAs in Escherichia coli.Mol Microbiol. 2011 Dec;82(6):1545-62. doi: 10.1111/j.1365-2958.2011.07907.x. Epub 2011 Nov 20. Mol Microbiol. 2011. PMID: 22040174 Free PMC article.
-
Hfq CLASH uncovers sRNA-target interaction networks linked to nutrient availability adaptation.Elife. 2020 May 1;9:e54655. doi: 10.7554/eLife.54655. Elife. 2020. PMID: 32356726 Free PMC article.
-
New aspects of RNA-based regulation by Hfq and its partner sRNAs.Curr Opin Microbiol. 2018 Apr;42:53-61. doi: 10.1016/j.mib.2017.10.014. Epub 2017 Nov 7. Curr Opin Microbiol. 2018. PMID: 29125938 Free PMC article. Review.
-
New molecular interactions broaden the functions of the RNA chaperone Hfq.Curr Genet. 2019 Dec;65(6):1313-1319. doi: 10.1007/s00294-019-00990-y. Epub 2019 May 18. Curr Genet. 2019. PMID: 31104083 Review.
Cited by
-
Extraribosomal Functions of Bacterial Ribosomal Proteins-An Update, 2023.Int J Mol Sci. 2024 Mar 3;25(5):2957. doi: 10.3390/ijms25052957. Int J Mol Sci. 2024. PMID: 38474204 Free PMC article. Review.
-
In vivo RNA interactome profiling reveals 3'UTR-processed small RNA targeting a central regulatory hub.Nat Commun. 2023 Dec 7;14(1):8106. doi: 10.1038/s41467-023-43632-1. Nat Commun. 2023. PMID: 38062076 Free PMC article.
References
-
- Adams PP, Flores Avile C, Popitsch N, Bilusic I, Schroeder R, Lybecker M, Jewett MW. In vivo expression technology and 5’ end mapping of the Borrelia burgdorferi transcriptome identify novel RNAs expressed during mammalian infection. Nucleic Acids Research. 2017;45:775–792. doi: 10.1093/nar/gkw1180. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

