Abstract
Expression of the Pseudomonas putida tod operon, which encodes enzymes for toluene metabolism, takes place from the PtodX promoter and is mediated by the TodS/TodT two component system. The sensor kinase TodS has a complex domain arrangement containing two functional modules, each harboring a sensor- and an autokinase domain and separated by a receiver domain. Based on site-directed mutagenesis of phosphoaccepting His-190, Asp-500, and His-760 and in vitro transphosphorylation experiments with recombinant TodS fragments, we show that TodS uses a multiple step phosphorelay mechanism to activate TodT. Toluene binding stimulates exclusively phosphorylation of His-190, which is followed by phosphotransfer to Asp-500 and subsequently to His-760 prior to phosphorylation of TodT Asp-57. Mutation of His-190, Asp-500, and H760A prevented up-regulation of toluene-mediated stimulation of TodT transphosphorylation in vitro and reduced in vivo expression of PtodX to the basal level. Calorimetric studies support that TodT binds to the C-terminal kinase module with a KD of ∼200 nm and 1:1 stoichiometry. This is the first report of a multiple step phosphorelay mechanism of a sensor kinase that involves two autokinase domains.
The adaptation of bacteria to changes in environmental signals is often mediated at the transcriptional level by two-component systems (TCS),2 which contain as basic components a sensor kinase and a response regulator (1). Sensor kinases generally recognize signal molecules, which leads to an alteration of their phosphorylation state and subsequently transphosphorylation of the corresponding cognate response regulator. The ratio of phosphorylated to nonphosphorylated response regulator directly influences promoter activity (2).
Based on domain arrangement and function, TCSs are grouped into two major families. One family, the so-called prototypal TCSs, is characterized by the fact that sensor kinases and response regulators each contain a single phosphorylatable site and phosphotransfer occurs directly from the sensor kinase to the response regulator. The other family comprises the multiple step phosphorelay systems (3), which in contrast to prototypal TCSs contain additional phosphorylation sites. These systems involve typically initial phosphorylation at the autokinase domain, followed by phosphotransfer to a receiver domain and subsequently to a phosphotransfer domain. The phosphoryl group for the transphosphorylation of the cognate response regulator departs from the latter domain (Fig. 1A). Multiple step phosphorelays have been described in which the phosphotransfer domain is an HPT domain (histidine-containing phosphotransfer, InterPro signature IPR008207, Ref. 4) or a Spo0B-like protein (IPR016120, Ref. 5). Both types of domains were shown to be devoid of autokinase activity (6, 7).
FIGURE 1.
A, schematic drawing of the domain arrangement of tripartite sensor kinases and TodS. Tm, transmembrane region; RRR, response regulator receiver domain. The domain arrangement for tripartite sensor kinases is taken from ArcB, the best studied family member, and the figure is adapted from Kwon et al. (8). The phosphorelay is indicated. The TodS domains were predicted by SMART (30). B, summary of truncated TodS versions used in this study. The ends of recombinant fragments were determined according to the domain annotation by SMART (30) and secondary structure prediction of TodS using the consensus method (38). Fragment ends were placed in regions for which a turn or coil secondary structure was predicted. All fragments were produced as His tag fusion proteins and purified as detailed for TodS in Lacal et al. (15). For further information see Table 1 and “Experimental Procedures.”
The initial phosphorylatable domains of a multiple step phosphorelay can either be part of a single protein, which is well documented for tripartite sensor kinases (Fig. 1) such as ArcB (8), BarA (9), EvgS, BvgS (10), and TorS (11), or on several proteins as exemplified for the KinA/Spo0F/Spo0B/Spo0A system in Bacillus subtilis (7).
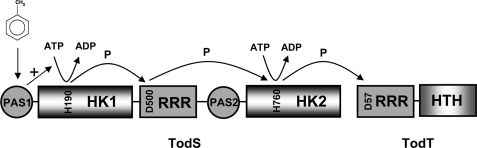
The domain organization of TodS from Pseudomonas putida DOT-T1E is different from that of the tripartite sensor kinases (Fig. 1). TodS has two modules each containing a PAS-type sensor domain and an autokinase domain. Because of this particular domain arrangement, members of this family have been termed “double sensor kinases.”
TodS forms, together with TodT, a TCS that regulates expression of the tod (toluene dioxygenase) operon in P. putida for toluene metabolism (12–16). Other members of the double sensor kinase family are also involved in the regulation of degradation pathways such as StyS (styrene degradation in P. fluorescence and Pseudomonas sp. Y2, Refs. 17 and 18), TutC (toluene degradation in Thauera aromatica, Ref. 19), or TmoS (toluene degradation in P. mendocina, Ref. 14). Furthermore, genome sequencing projects have revealed that TodS-like proteins are found in other hydrocarbon-degrading bacteria such as Dechloromonas aromatica (20) and Methylibium petroleiphilum (21). The reason for this apparent functional link between the domain arrangement of TodS-type sensor kinases, and their involvement in the regulation of hydrocarbon degradation is unknown.
The 108-kDa TodS protein is among the largest sensor kinases ever described. We developed a protocol for the purification of the full-length protein and showed that it exhibits a broad effector specificity, recognizing a wide variety of mono- and biaromatic compounds (15, 16). We have also shown that effector molecules bind to the N-terminal PAS domain, causing an increase in the amount of phosphorylated TodS, which in turn stimulates transphosphorylation of TodT (15, 16). TodT binds to three different sites at the PtodX promoter in a cooperative and hierarchical manner. TodT-P in conjunction with IHF (integration host factor) was found to stimulate promoter activity (22, 23). However, the functional reason for the complex architecture of TodS remains unknown and is the focus of this work. Two different modes of activation can be envisaged: either each of the modules containing a PAS and a kinase domain are independent units responding to different signals and serving potentially different regulators, or alternatively, all three phosphorylation sites are functionally linked by intramolecular communication. Using a set of site-directed mutants as well as different recombinant fragments, we were able to establish that TodS uses also a multiple step phosphorelay mechanism involving amino acids His-190→Asp-500→His-760 followed by phosphorylation of Asp-57 at TodT.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids—The bacterial strains and plasmids used in this study are listed in Table 1. Cells were grown on LB medium supplemented with appropriate antibiotics.
TABLE 1.
Bacterial strains and plasmids used in this study
| Relevant characteristicsa | Refs. | |
|---|---|---|
| E. coli DH5αF′ | F'/hsdR17, recA1, gyrA | (39) |
| E. coli BL21(DE3) | F-, ompl, hsdSB (r-B m-B) | (40) |
| P. putida DOT-T1E | phototroph, Tol+ (tod pathway) | (41) |
| P. putida DOT-T1EtodST | DOT-T1E, todST::Km, Tol-, containing pMlR66, GmR | (14) |
| pET-28b(+) | KmR, protein expression vector | (Novagen) |
| pMIR77 | TcR, PtodX::'lacZ inserted in pMP220 | (14) |
| pMIR66 | GmR, containing the todST genes inserted in pBBR1MCS-5 | (14) |
| pTodS | pET28b (+) containing todS gene | (15) |
| pNTodS | pET28b (+) containing todS sequence 1-1752 (amino acids 1-584, NTodS-long) | (15) |
| pCTodS | pET28b (+) containing todS sequence 1356-2934 (amino acids 452-978, CTodS-long) | (15) |
| pHK1 | pET28b (+) containing todS sequence 49-1347 (amino acids 16-449, NTodS-short) | This work |
| pRRR | pET28b (+) containing todS sequence 1348-1755 (amino acids 450-585, MTodS) | This work |
| pHK2 | pET28b (+) containing todS sequence 1756-2928 (amino acid 586-976, CTodS-short) | This work |
| pAB1 | pTodS containing the mutation H190A | This work |
| pAB2 | pTodS containing the mutation D500A | This work |
| pAB3 | pTodS containing the mutation H760A | This work |
| pAB4 | pMIR66 containing the mutation H190A | This work |
| pAB5 | pMIR66 containing the mutation D500A | This work |
| pAB6 | pMIR66 containing the mutation H760A | This work |
| pJLTodT | pET28b containing the todT gene | (15) |
| pTodTD57A | pJLTodT containing the mutation D57A | (15) |
Abbreviations: GmR, KmR,TcR resistance to gentamicin, kanamycin, and tetracycline, respectively. Tol+/Tol- indicates that the strain grows or fails to grow on toluene, respectively.
Recombinant DNA Techniques—Site-directed mutations in the todS gene were introduced into plasmids pTodS (for protein expression; see Table 1) and pMIR66 (for β-galactosidase measurements; see Table 1) using the QuikChange mutagenesis kit (Stratagene) according to the manufacturer's instructions. Oligonucleotides used are listed in Table 2. For the generation of truncated versions of TodS the corresponding gene fragments were amplified with appropriate primers using Taq Polymerase (Eppendorf) and pMIR66 as the template. The resulting products were digested with NheI and BamHI and cloned into pET28b(+) (Novagen) linearized with the same enzymes. All plasmids were verified by sequencing the insert and flanking regions. Plasmids pHK1, pRRR, and pHK2 express the N-terminal module, the receiver domain and the C-terminal module of the TodS protein, respectively, and the corresponding truncated proteins produced from these constructs were called NTodS-short, MTodS, and CTodS-short, respectively.
TABLE 2.
Oligonucleotides used to generate recombinant TodS fragments and site-directed mutants
| Plasmids | Sequence (5′-3′) | |
|---|---|---|
| For expression plasmids of TodS fragments | ||
| pHK1 | GGGGCTAGCAATTATTATAATATCTGCCTCAAG | |
| CGCGGATCCTCAGTCGGATTGAAATGGAAG | ||
| pRRR | GAGGCTAGCCAGCCTCGGGTGCTAAT | |
| CACGGATCCTCACCCCTGATCGGAAAGCTC | ||
| pHK2 | GCATACGGATCCTCACTGTCTGGCAGGGATACT | |
| For site-directed mutagenesis of plasmids pTodS and pMIR66 | ||
| pAB1 and pAB4 | TTGCCAAGGTGAGCGCTGAGTTGCGCACTCCa | |
| GGAGTGCGCAACTCAGCGCTCACCTTGGCAAa | ||
| pAB2 and pAB5 | ACCTGTTGATTACAGCCCTGATGATGCCTa | |
| AGGCATCATCAGGGCTGTAATCAACAGGTa | ||
| pAB3 and pAB6 | GCAGCCTATATTGCAGCCGAGATAAACCAACCGa | |
| CGGTTGGTTTATCTCGGCTGCAATATAGGCTGCa | ||
Mismatch nucleotides for amino acid replacement to alanine are underlined.
Overexpression and Purification of Proteins—TodS, its site-directed mutants and recombinant fragments were expressed and purified as detailed for TodS in Lacal et al. (15). TodT and TodTD57A were purified as described for TodT in the same publication.
Phosphorylation Assays—Prior to analysis, all proteins were dialyzed into 50 mm Tris-HCl, pH 7.5, 200 mm KCl, 2 mm MgCl2, 0.1 mm EDTA, 10% (v/v) glycerol, and 2 mm dithiothreitol.
Autophosphorylation of TodS, its recombinant fragments, or site-directed mutants was carried out with 200 μm ATP containing 4 μCi of [γ-32P]ATP. To determine the effect of toluene on autophosphorylation a 2-mm stock solution of toluene was prepared in dialysis buffer and added to the sample to a final concentration of 100 μm. The corresponding volume of buffer was added to the control sample. After different time intervals, samples were removed, and the reaction was stopped by the addition of ice-cold 6× Laemmli loading buffer or flash-freezing prior to analysis on SDS-PAGE.
For transphosphorylation assays between TodS and TodT or TodTD57A, 20 μm TodS was incubated for 20 min with 200 μm ATP containing 4μCi [γ-32P]ATP, then a slight molar excess of TodT was added. Reactions were stopped after 15 min.
For transphosphorylation experiments between individual fragments of TodS, NTodS- at 60 μm was incubated with 200 μm ATP containing 4μCi [γ-32P]ATP for 20 min. ATP was then separated from the protein by gel filtration on NAP-5 columns (GE Healthcare). Phosphorylated protein was then incubated with equimolar amounts of MTodS, and samples were removed at different time intervals.
For transphosphorylation assays between recombinant TodS fragments and TodT the different TodS polypeptides at 20 μm where first incubated in 200 μm ATP containing 4 μCi [γ-32P]ATP for 20 min. Subsequently MTodS or an equivalent volume of buffer was added to all samples. After another 10 min of incubation, an equimolar amount of TodT was added. Samples were taken after 10 min for analysis by SDS-PAGE.
Quantification of 32P-Labeled Proteins—Gels were dried under a vacuum at 80 °C and exposed to a phosphorimager (Personal FX equipment, Bio-Rad). Densitometric analysis was done with QuantityOne® software.
Isothermal Titration Calorimetry (ITC)—Freshly purified TodS and TodT were dialyzed into 40 mm Tris-HCl, pH 7.3, 200 mm KCl, 300 mm imidazole, 10 mm MgCl2, 1 mm dithiothreitol, and 10% (v/v) glycerol. Experiments were carried out at 25 °C using a VP-microcalorimeter (MicroCal, Northampton, MA). TodS at a concentration of 1.6 μm was titrated with aliquots of 23 μm TodT. This experiment was repeated using TodS and TodT solutions to which 1 mm freshly prepared ATP was added. Injection of 23 μm TodT into dialysis buffer resulted in small and uniform peaks typical of dilution heats. Data were analyzed with the MicroCal version of ORIGIN using the one binding site model.
β-Galactosidase Assays—To quantify the expression from the TOD pathway promoter we used P. putida DOT-T1E todST, which is a knock-out mutant for todST, bearing pMIR77 (PtodX::lacZ) and pMIR66 or derivatives thereof (Table 1). Fresh LB medium was inoculated with a single colony from LB agar plates containing the appropriate antibiotics and cultured at 30 °C overnight. These cultures were diluted 100-fold in the same medium supplemented or not with 1.5 mm toluene, and cell growth was monitored over time. When the cultures reached a turbidity of 0.8 at 660 nm β-galactosidase activity was determined in permeabilized whole cells as described by Miller (24).
RESULTS
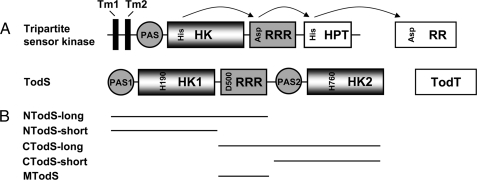
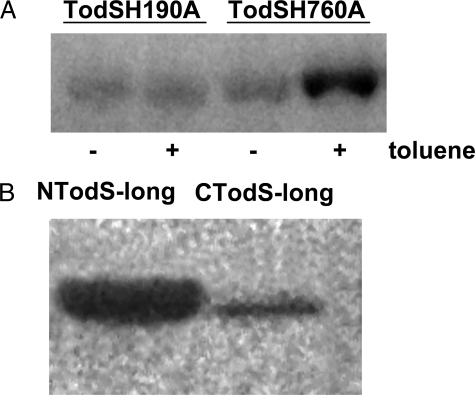
TodS Contains Two Functionally Active Histidine Autokinase Domains—TodS was predicted to contain two autokinase domains (Fig. 1), and initial experiments were aimed at verifying whether both domains posses autophosphorylation activity. To this end, alanine substitution mutants of the histidine residues predicted to be the phosphoacceptors in each autokinase domain, namely His-190 and His-760, were generated by site-specific mutagenesis, and autophosphorylation assays in the absence and presence of toluene were carried out. Equal amounts of TodSH190A and TodSH760A were incubated with [γ-32P]ATP for 20 min, and samples were analyzed by SDS-PAGE. In the absence of toluene (Fig. 2A), a band of similar intensity was detected for each mutant. These results were corroborated by autophosphorylation assays of the recombinant fragments NTodS-long and CTodS-long (Fig. 1B), each of which contain a single autokinase domain and a receiver domain. As expected, both proteins were phosphorylated (Fig. 2B), although the amount of phosphorylated CTodS-long was less than NTodS-long. These results show that TodS possesses two functionally active kinase domains with similar basal activity.
FIGURE 2.
Autophosphorylation of site-directed mutants and recombinant fragments of TodS. Protein at 20 μm was incubated with 200 μm ATP (containing 4 μCi of [γ-32P]ATP) for 20 min prior to analysis by SDS-PAGE. A, autophosphorylation of TodSH190A and TodSH760A in the presence and absence of 100 μm toluene. Both mutants have similar basal activities but toluene stimulates only TodSH760A. B, autophosphorylation of NTodS-long and CTodS-long in the absence of toluene.
Toluene Binding to the N-terminal PAS Domain Increases Phosphorylation of Its Neighboring Autokinase Domain—We have shown previously that effector molecules such as toluene bind exclusively to the N-terminal PAS domain of TodS (Figs. 1, 15, 16). Because this sensor kinase has two functional autokinase domains, effector binding could stimulate either one of the domains or potentially both. To address this question, autophosphorylation assays of mutants TodSH190A and TodSH760A were repeated in the presence of toluene. No alteration in the phosphorylation state as compared with the absence of toluene was observed for mutant TodSH190A, whereas a significant increase in phosphorylation in response to toluene was noted for mutant TodSH760A (Fig. 2A). Because phosphorylation of the latter mutant is solely due to the action of the N-terminal kinase domain, our data indicate that toluene increased the phosphorylation level of only the N-terminal kinase domain.
Evidence for an Intramolecular Phosphorelay at TodS—TodS is predicted to contain three phosphorylatable amino acids: His-190 and His-760 of the autokinase domains and Asp-500 of the response regulator receiver domain (Fig. 1). It has been reported that recombinant domains of large proteins can associate in a native-like fashion without significant loss of protein activity when mixed in vitro (25). To provide proof for phosphotransfer between the different domains of TodS, we generated recombinant proteins corresponding to the NTodS-short, MTodS, and CTodS-short fragments (Fig. 1B), each of which contained a single phosphoaccepting residue (His-190, Asp-500, and His-760, respectively). These three recombinant proteins with a single phosphoaccepting residue were subjected to autophosphorylation assays with ATP.
The data available to date on the response regulator receiver domain indicate that it cannot be phosphorylated by ATP. To verify whether this also holds for TodS, the recombinant fragment MTodS (Fig. 1B) was subjected to phosphorylation assays with [γ-32P]ATP, which unequivocally revealed that it was not phosphorylated by ATP (data not shown).
Satisfactory autophosphorylation was observed for NTodS-short, but CTodS-short was devoid of activity (data not shown). Therefore, attempts were focused on detecting transphosphorylation between NTodS-short and MTodS, containing His-190 and Asp-500, respectively. To this end, NTodS-short was phosphorylated in the presence of toluene and [γ-32P]ATP and applied to a NAP-5 gel filtration column to separate the protein from ATP. A stoichiometric amount of MTodS was added to the resulting protein, and samples were taken at different times for SDS-PAGE analysis. As shown in Fig. 3, the band corresponding to NTodS-short gradually disappeared whereas a band representing phosphorylated MTodS became visible. Maximum MTodS phosphorylation was seen after 10 min, before this protein gradually dephosphorylated. This strongly suggests the existence of intramolecular phosphotransfer in TodS.
FIGURE 3.
Transphosphorylation between recombinant fragments of TodS. NTodS was phosphorylated with 200 μm ATP (containing 4 μCi of [γ-32P]ATP), and the solution was applied to NAP-5 gel filtration columns (GE Healthcare) to separate protein from ATP. An equimolar amount of MTodS was added to the protein fraction. At the time intervals indicated, samples were removed for SDS-PAGE analysis.
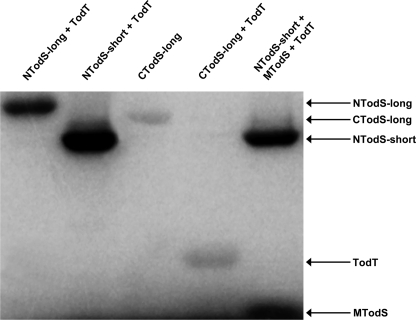
TodT Transphosphorylation Occurs from the C-terminal Kinase Domain of TodS—Because TodS contains two kinase domains, transphosphorylation to TodT may occur from either of the domains or from both. To cast light on this issue, transphosphorylation between various TodS fragments and TodT was studied (Fig. 4). To this end TodS polypeptides were phosphorylated for 20 min, which was followed by the addition of an equimolar amount of TodT. The first two lanes in Fig. 4 illustrate that NTodS-short and NTodS-long (see Fig. 1) are efficiently phosphorylated by [γ-32P]ATP. However, no transphosphorylation to TodT was detectable. Lane 3 shows that CTodS-long was phosphorylated, although to a lower extent than NTodS as already noted in Fig. 2. However, the addition of TodT to CTodS-long as shown in lane 4 of Fig. 4 resulted in a quantitative transfer of the phosphoryl groups to TodT. The last lane, representing a mixture of NTodS-short, MTodS and TodT, was another control experiment which illustrated that NTodS-short maintained its capacity to transphosphorylate MTodS, but failed to phosphorylate TodT. In summary, these data are consistent with phosphotransfer from the C-terminal kinase domain toward TodT.
FIGURE 4.
Transphosphorylation between recombinant fragments of TodS and TodT. TodS fragments NTodS-short, N-TodS-long, and CTodS-long at 20 μm where incubated with 200 μm ATP containing 4 μCi of [γ-32P]ATP for 20 min. An equimolar amount of MTodS was added to the sample in lane 5, and the corresponding amount of buffer to the remaining samples. After 10 min of incubation, an equimolar amount of TodT was added to the samples in lanes 1, 2, 4, and 5, and the corresponding amount of buffer was added to the sample in lane 3. Samples were taken after another 10 min of incubation.
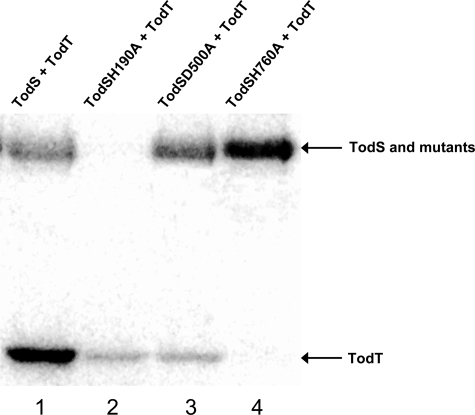
Analysis of Site-directed TodS Mutants Confirms the Multiple Step Phosphorelay Mechanism and TodT Transphosphorylation from His-760—To provide further support for the existence of a His-190→Asp-500→His-760 phosphorelay, transphosphorylation assays were conducted with TodT and the three TodS mutants in which the phosphoaccepting amino acid was replaced with alanine. Equal amounts of TodS, TodSH190A, TodSD500A, and TodSH760A were incubated with [γ-32P]ATP for 20 min before TodT was added. Experiments were carried out in the presence of toluene which implies that activity of HK1 domain is stimulated whereas HK2 domain operates at basal activity (Fig. 2A). Transphosphorylation assays of the wild-type protein showed bands for both TodS and TodT (Fig. 5, lane 1). The band corresponding to TodT dominated, indicating that transphosphorylation was a relatively rapid process, which is in agreement with studies of other TCSs (26).
FIGURE 5.
Transphosphorylation between TodT and TodS or its mutants TodSH190A, TodSD500A, and TodSH760A. Wild-type and mutant TodS at 20 μm were incubated with 200 μm ATP (containing 4μCi of [γ-32P]ATP) and 100 μm toluene for 20 min. An equimolar amount of TodT was added, and samples were removed for SDS-PAGE analysis after 15 min of incubation.
However, transphosphorylation assays of TodSH760A showed only a single strong band at the TodS level (Fig. 5, lane 4), whereas no band was seen for TodT. The absence of any transphosphorylation demonstrates that the phosphoryl group departed from residue His-760 in TodS toward TodT, which confirms data presented in Fig. 4. The strong band corresponding to TodSH760A confirms that toluene stimulated phosphorylation, but in the absence of transphosphorylation the phosphoryl group apparently remained at His-190 and Asp-500. Most interestingly, the amount of phosphorylated TodT seen in the transphosphorylation of TodSH190A (lane 2) and TodSD500A (lane 3) was very similar, probably because of the weak basal activity of the C-terminal kinase domain. Previous experiments have shown that toluene stimulated autophosphorylation of TodSD500A (data not shown) but not of TodSH190A (Fig. 2). However, this stimulation was not reflected as an increase in phosphorylated TodT in transphosphorylation assays (Fig. 5). These findings indicate that Asp-500 was essential to transmit an increase in phosphorylation of the N-terminal kinase domain to the C-terminal kinase domain. The notion that the His-190→Asp-500→His-760 phosphorelay is interrupted by replacing Asp-500 was supported by the fact that the amount of phosphorylated TodSD500A was larger than the phosphorylated wild-type TodS.
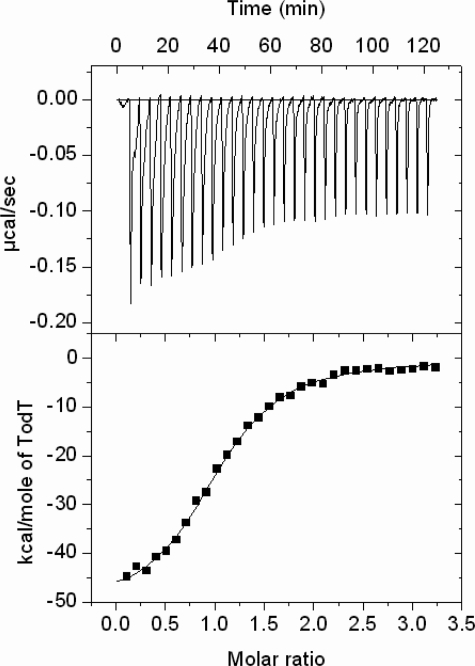
TodT Binds TodS with 1:1 Stoichiometry: The Presence of ATP Is Essential for Interaction—We used ITC (27) to study the interaction between TodS and TodT. ITC experiments of TodS with TodT at three different temperatures did not reveal binding heats, which suggested, unexpectedly, that the two proteins did not interact. Another series of ITC titrations of TodS with different nucleotides such as ATP, ADP, and the non-hydrolyzable ATP analogue AMP-PNP revealed, surprisingly, that TodS bound only ATP. We then studied whether TodS interacted with TodT in the presence of 1 mm ATP, by adding a freshly prepared solution to both proteins prior to the experiment. The raw data from this experiment (Fig. 6) showed that initial peaks were broad at their base, which could be explained by the fact that initial binding heats were followed by heats from the kinetically slower process of TodS-TodT transphosphorylation (note: binding processes that are not followed by a kinetically slow event give rise to peaks which are narrow at its base). This distorted the enthalpic and entropic contribution, but did not impact on the binding constant or the stoichiometry of the interaction (27). After each injection the signal returned to the baseline, which enabled correct peak integration and curve fitting (Fig. 6, lower panel). Data analysis revealed that both proteins interacted with a stoichiometry of 1.06 ± 0.1 and a dissociation constant of 214 ± 10 nm. The control experiment with TodT/ATP and buffer/ATP yielded small, uniform peaks. When this experiment was repeated in the presence of 1 mm ADP, AMP-PNP, or 100 μm toluene, no interactions was observed between TodS and TodT.
FIGURE 6.
ITC study of the interaction of TodS with TodT. Upper panel, raw titration data of 1.6 μm TodS with aliquots of 23 μm TodT in the presence of 1 mm ATP. For further experimental details see “Experimental Procedures” and Krell (27). Lower panel, integrated and dilution-corrected peak areas from the raw data. The curves were fitted with the “one-binding-site” model of the MicroCal version of ORIGIN.
Subsequently the interaction between NTodS-long and CTodS-long with TodT was studied in an analogous fashion, i.e. in presence of 1 mm ATP. CTodS-long was found to bind to TodT with an affinity of 65 ± 7nm (data not shown). No binding heats were observed for the titration of NTodS-long with TodT.
TodT Asp-57 Is the Last Step in the TodS/TodT Phosphorylation Cascade—The data presented so far suggested the existence of an intramolecular phosphorylation cascade at TodS, and that residue His-760 of TodS is the amino acid that donates the phosphoryl group to the response regulator. Previous in vitro transcription studies (15) suggested that Asp-57 operates as the phosphoaccepting residue. To verify this prediction transphosphorylation assays between TodS and TodT or TodTD57A were carried out (Fig. 7). No transphosphorylation was detected for the TodTD57A mutant, which indicated that this amino acid is the last step in the His-190→Asp-500→His-760→Asp-57 phosphorelay in the TodS/TodT TCS.
FIGURE 7.
Transphosphorylation between TodT or TodTD57A and TodS. 20 μm TodS was incubated with 200 μm ATP (containing 4 μCi of [γ-32P]ATP) and 100 μm toluene for 20 min. TodT or TodTD57A was then added to a final concentration of 30 μm, and cultures were incubated for 15 min and analyzed by SDS-PAGE.
In Vivo Proof for the Deduced Phosphorylation Cascade in the TodS/TodT System—The effect of the amino acid substitutions H190A, D500A, and H760A was evaluated in vivo by measuring expression from PtodX promoter fused to `lacZ. Plasmid pMIR77 bearing the PtodX::lacZ fusion was transformed in the knock-out mutant P. putida DOT-T1EtodST– bearing or not pMIR66 (todST) or plasmids pAB4, pAB5, and pAB6, which are pMIR66 derivative with a todS mutant allele that produces TodSH190A, TodSD500A, and TodSH760A, respectively. In cells with pMIR77 plus pMIR66, expression from PtodX was basal and low (25–40 Miller units) in the absence of toluene, and increased to 5000 ± 500 Miller units when toluene was added. However, in the three strains bearing pAB4, pAB5, and pAB6 encoding the above-mentioned three TodS mutants, expression from PtodX was below 30 Miller units regardless of the presence of toluene. Therefore, the mutation of His-190, Asp-500, and His-760 entirely abrogates toluene-mediated induction of PtodX activity in vivo. These data entirely support (a) that toluene stimulates phosphorylation of H190, (b) that Asp-500 is essential for intra-TodS phosphotransfer and that there is no direct link between the two kinase domains, and (c) that His-760 is the site for transphosphorylation of TodT.
DISCUSSION
TodS Belongs to a New Family of Multiple Step Phosphorelay Sensor Kinases—Two-component systems contain as basic components a sensor kinase and a response regulator. Data currently available indicate that response regulators involved in transcriptional control form a relatively well-conserved family of proteins. Galperin (28) showed that over 80% of DNA binding response regulators belong to either the NarL or OmpR families, which are characterized by an N-terminal receiver domain and a C-terminal helix-turn-helix motif-containing DNA binding domain. In contrast to the relative conservation of response regulators is the extreme functional and structural diversity of sensor kinases (29), which can be membrane-bound or not, have a large variety of different sensor domains located either in the periplasm or cytosol and recognize signals in many different ways (29). To fully comprehend the physiological relevance of TCSs it appears indispensable to explore the functional and structural diversity of sensor kinases.
TodS is clearly different from other sensor kinases characterized to date because it contains two modules each of which has a sensor and a kinase domain. Based on our data, we propose the functional model of TodS depicted in Fig. 8. Initial binding of toluene increases the phosphorylation state of the N-terminal autokinase domain. The phosphoryl group is subsequently transferred to the internal receiver domain, and eventually to the C-terminal kinase domain before it is transferred to Asp-57 of TodT, which is the primary regulator of the PtodX promoter.
FIGURE 8.
Mechanistic model for the function of TodS. The binding of effectors, such as toluene, occurs at the N-terminal PAS sensor domain, which stimulates phosphorylation of its neighboring kinase domain. The phosphoryl group is then passed on to Asp-500 of the internal response regulator receiver domain and subsequently to His-760 of the C-terminal kinase domain, which is the site for transphosphorylation of TodT. The C-terminal kinase domain has autokinase activity, and it remains to be elucidated whether this is modulated by potential recognition of a signal by the PAS 2 sensor domain.
Effector binding stimulated phosphorylation of His-190, whereas TodT phosphorylation occurred from His-760, evidence of communication between the two kinase domains. Available data support the absence of a direct communication between the two kinase domains, and that Asp-500 is essential for establishing this link. This statement is based on two lines of evidence. First, in vitro autophosphorylation studies show that toluene stimulates activity of TodSD500A (increased phosphorylation at His-190) but not that of the H190A mutant. However, the amount of phosphorylated TodT in transphosphorylation experiments with TodSD500A and TodSH190A is similar (Fig. 5). Second, no toluene-mediated stimulation of transcription from PtodX was observed in in vivo gene expression studies of mutant D500A. At present we have not explored whether the phosphorelay occurs in the inverse direction (His-760→Asp-500→His-190), which could be of functional and physiological importance.
The phosphorelay described for TodS is in several aspects comparable to that of previously described systems such as the ArcB system. In both cases the sequence of transphosphorylation involves the amino acids H1-D-H2. In vivo data for ArcB show that there is no direct transphosphorylation between H1 and H2 and that the response regulator ArcA is exclusively phosphorylated by H2 (8). The data presented here indicate that the TodS phosphorelay shares the same characteristics. However, the TodS phosphorelay distinguishes itself from the ArcB system in the fact that it employs a second autokinase domain, instead of an HPT domain, as phosphotransfer domain and that it also harbors an additional PAS domain. Both differences will be discussed in some more detail below.
Sequence analysis of TodS using SMART (30) identifies with high confidence two histidine autokinase domains (defined by signatures IPR003661 and IPR003594), which were termed HK1 and HK2. Here we show that both domains possess autokinase activity using ATP as cofactor. In general, the biological function of a histidine autokinase domain consists in its autophosphorylation and the subsequent catalysis of phosphotransfer to either the cognate response regulator or, in the case of phosphorelays, to an internal receiver domain. In the case of TodS, the HK1 domain fulfills this function. However, the phosphotransfer domain, typically HPT domains or Spo0B-like proteins in other phosphorelays, is in the case of TodS another histidine autokinase domain. The fact that a histidine kinase domain operates as a phosphotransfer domain in a phosphorelay corresponds thus to a new biological function for this domain. HK1 and HK2 share only a modest degree of sequence identity of 23%. Furthermore both domains belong to different classes (HPK1a and HPK4, respectively, for HK1 and HK2) according to the sequence-based classification proposed by Grebe and Stock (31). This indicates that TodS might have evolved as result of a gene fusion rather than gene duplication event.
The obvious question is thus why TodS has an autokinase phosphotransfer domain whereas the large majority of other phosphorelay systems employ an HPT domain instead. The answer to that question may lie in the second feature, which distinguishes TodS from other known phosphorelay systems, which is the presence of an additional PAS domain located in sequence just upstream the HK2 domain. In general, PAS domains are involved in sensing of small molecules (32), and it is therefore likely that PAS 2 of TodS also exerts this function. We have shown that TodS His-760 can receive phosphoryl groups from either ATP or Asp-500. In this context it appears possible that either or both activities are modulated by an as yet unknown signal, which is recognized by the C-terminal PAS sensor domain. We have shown previously that aromatic effector molecules bind exclusively to the N-terminal PAS domain. The notion that both PAS domains might respond to different signals is underlined by the weak sequence identity of 17% between both PAS domains. For the TodS homologue StyS it was proposed that the C-terminal PAS domain might respond to the cell redox potential (33). This would also agree with the concept that the physiological reason for complex multidomain sensor kinases lies in their capacity to respond to different signals enabling for a fine-tuning of the transcriptional response. The existence and mechanism of action of a secondary signal, which might modulate TodS activity remain to be documented.
Sensor kinases predicted to contain two autokinase domains separated in sequence by a receiver domain are present in numerous bacteria such as Legionella pneumophila, Bradyrhizobium sp., Stigmatella aurantica, and Anabaena variabilis or eukaryotes like Dictyostelium discoideum or Filobasidiella neoformans. It is thus possible that these proteins also use a multiple step phosphorelay mechanism.
Requirement of ATP for TodS/TodT Interaction—There is no doubt that the TodS/TodT system represents a novel regulatory mechanism and presents specific features not found in other two component systems. A feature which clearly distinguishes TodS/TodT from other systems concerns the molecular interaction between the two proteins. A large series of studies on other TCSs converged on the finding that both components interact in vitro with relatively modest affinity for a protein-protein interaction, as exemplified by FixL/FixJ (KD = 4 μm, Ref. 34), EnvZ/OmpR (1.2 μm, Ref. 35), CheA/CheY (0.9–1.3 μm, Ref. 36), and EvgS/EvgA (1.2 μm, Ref. 37). In all cases, the presence of ATP was not required for molecular interaction. ITC experiments of TodS with TodT at three different temperatures did not reveal binding heats, a finding which strongly suggests that the two proteins do not interact. The same result was obtained in the presence of 100 μm toluene. However, when the experiments were repeated with both protein solutions containing 1 mm ATP, a sigmoid binding curve was observed (Fig. 6). We hypothesize that ATP binding at TodS induces conformational changes enabling interaction with TodT. The cellular ATP concentration is estimated to be between 3 and 5 mm implying TodS saturation, and the potential physiological relevance of this finding awaits further study.
Acknowledgments
We thank Carmen Lorente and M. Mar Fandila for secretarial assistance, Cristina García Fontana for technical assistance as well as K. Shashok for improving the English in this manuscript.
This work was supported by grants from the Spanish Ministry of Science and Education (CICYT BIO-2006-05668, BFU 2005-0487-C02-02, CSD 2007-00010) and Grants CIV344 and CIV1912 from the Junta de Andalucía (to J. L. R. and T. K.), respectively.
Footnotes
The abbreviations used are: TCS, two-component system; HK, histidine kinase; HPT, histidine-containing phosphotransfer domain; ITC, isothermal titration calorimetry; PAS domain, Per-Arnt-Sim domain; AMP-PNP, adenosine 5′-(β,γ-imino)triphosphate.
References
- 1.Ulrich, L. E., Koonin, E. V., and Zhulin, I. B. (2005) Trends Microbiol. 13 52–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stock, A. M., Robinson, V. L., and Goudreau, P. N. (2000) Annu. Rev. Biochem. 69 183–215 [DOI] [PubMed] [Google Scholar]
- 3.Appleby, J. L., Parkinson, J. S., and Bourret, R. B. (1996) Cell 86 845–848 [DOI] [PubMed] [Google Scholar]
- 4.Zhang, W., and Shi, L. (2005) Microbiology 151 2159–2173 [DOI] [PubMed] [Google Scholar]
- 5.Varughese, K. I., Madhusudan Zhou, X. Z., Whiteley, J. M., and Hoch, J. A. (1998) Mol. Cell. 2 485–493 [DOI] [PubMed] [Google Scholar]
- 6.Georgelis, D., Lynch, A. S., and Lin, E. C. (1997) J. Bacteriol. 179 5429–5435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burbulys, D., Trach, K. A., and Hoch, J. A. (1991) Cell 64 545–552 [DOI] [PubMed] [Google Scholar]
- 8.Kwon, O., Georgellis, D., and Lin, E. C. (2000) J. Bacteriol. 182 3858–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tomenius, H., Pernestig, A. K., Méndez-Catalá, C. F., Georgellis, D., Normark, S., and Melefors, O. (2005) J. Bacteriol. 187 7317–7324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perraud, A. L., Kimmel, B., Weiss, V., and Gross, R. (1998) Mol. Microbiol. 27 875–887 [DOI] [PubMed] [Google Scholar]
- 11.Jourlin, C., Ansaldi, M., and Méjean, V. (1997) J. Mol. Biol. 267 770–777 [DOI] [PubMed] [Google Scholar]
- 12.Lau, P. C. K., Wang, Y., Patel, A., Labbé, D., Bergeron, H., Brousseau, R., Konishi, Y., and Rawlings, M. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 1453–1458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mosqueda, G., Ramos-González, M. I., and Ramos, J. L. (1999) Gene 232 69–76 [DOI] [PubMed] [Google Scholar]
- 14.Ramos-González, M. I., Olson, M., Gatenby, A. A., Mosqueda, G., Manzanera, M., Campos, M. J., Vílchez, S., and Ramos, J. L. (2002) J. Bacteriol. 184 7062–7067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lacal, J., Busch, A., Guazzaroni, M. E., Krell, T., and Ramos, J. L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 8191–8196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Busch, A., Lacal, J., Martos, A., Ramos, J. L., and Krell, T. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 13774–13779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leoni, L., Ascenzi, P., Bocedi, A., Rampioni, G., Castellini, L., and Zennaro, E. (2003) Biochem. Biophys. Res. Commun. 303 926–931 [DOI] [PubMed] [Google Scholar]
- 18.Velasco, A., Alonso, S., García, J. L., Perera, J., and Díaz, E. (1998) J. Bacteriol. 180 1063–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coschigano, P. W., and Young, L. Y. (1997) Appl. Environ. Microbiol. 63 652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coates, J. D., Chakraborty, R., Lack, J. G., O'Connor, S. M., Cole, K. A., Bender, K. S., and Achenbach, L. A. (2001) Nature 411 1039–1043 [DOI] [PubMed] [Google Scholar]
- 21.Kane, S. R., Chakicherla, A. Y., Chain, P. S., Schmidt, R., Shin, M. W., Legler, T. C., Scow, K. M., Larimer, F. W., Lucas, S. M., Richardson, P. M., and Hristova, K. R. (2007) J. Bacteriol. 189 1931–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lacal, J., Guazzaroni, M. E., Busch, A., Krell, T., and Ramos, J. L. (2008) J. Mol. Biol. 376 325–337 [DOI] [PubMed] [Google Scholar]
- 23.Lacal, J., Guazzaroni, M. E., Gutiérrez-del-Arroyo, P., Busch, A., Vélez, M., Krell, T., and Ramos, J. L. (2008) J. Mol. Biol. 384 1037–1047 [DOI] [PubMed] [Google Scholar]
- 24.Miller, J. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- 25.Krell, T., Renauld-Mongénie, G., Nicolaï, M. C., Fraysse, S., Chevalier, M., Bérard, Y., Oakhill, J., Evans, R. W., Gorringe, A., and Lissolo, L. (2003) J. Biol. Chem. 278 14712–14722 [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto, K., Hirao, K., Oshima, T., Aiba, H., Utsumi, R., and Ishihama, A. (2005) J. Biol. Chem. 280 1448–1456 [DOI] [PubMed] [Google Scholar]
- 27.Krell, T. (2008) Microb. Biotechnol. 1 126–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galperin, M. Y. (2006) J. Bacteriol. 188 4169–4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascher, T., Helmann, J. D., and Unden, G. (2006) Microbiol. Mol. Biol. Rev. 70 910–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schultz, J., Milpetz, F., Bork, P., and Ponting, C. P. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grebe, W., and Stock, J. B. (1999) Adv. Micro. Physiol. 41 139–227 [DOI] [PubMed] [Google Scholar]
- 32.Taylor, B. L., and Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. 63 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rampioni, G., Leoni, L., Pietrangeli, B., and Zennaro, E. (2008) BMC Microbiol. 8 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sousa, E. H., Gonzalez, G., and Gilles-Gonzalez, M. A. (2005) Biochemistry 44 15359–15365 [DOI] [PubMed] [Google Scholar]
- 35.Cai, S. J., and Inouye, M. (2002) J. Biol. Chem. 277 24155–24161 [DOI] [PubMed] [Google Scholar]
- 36.Stewart, R. C., and van Bruggen, R. (2004) Biochemistry 43 8766–8777 [DOI] [PubMed] [Google Scholar]
- 37.Perraud, A. L., Rippe, K., Bantscheff, M., Glocker, M., Lucassen, M., Jung, K., Sebald, W., Weiss, V., and Gross, R. (2000) Biochim. Biophys. Acta 1478 341–354 [DOI] [PubMed] [Google Scholar]
- 38.Deleage, G., Blanchet, C., and Geourjon, C. (1997) Biochimie (Paris) 79 681–686 [DOI] [PubMed] [Google Scholar]
- 39.Raleigh, E. A., Trimarchi, R., and Revel, H. (1989) Genetics 122 279–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier, F. W., Rosenberg, A. H., Dunn, J. J., and Dubendorff, J. W. (1990) Methods Enzymol. 185 60–89 [DOI] [PubMed] [Google Scholar]
- 41.Ramos, J. L., Duque, E., Huertas, M. J., and Haïdour, A. (1995) J. Bacteriol. 177 3911–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]