Summary
The recent emergence of the novel, pathogenic SARS-coronavirus 2 (SARS-CoV-2) in China and its rapid national and international spread pose a global health emergency. Cell entry of coronaviruses depends on binding of the viral spike (S) proteins to cellular receptors and on S protein priming by host cell proteases. Unravelling which cellular factors are used by SARS-CoV-2 for entry might provide insights into viral transmission and reveal therapeutic targets. Here, we demonstrate that SARS-CoV-2 uses the SARS-CoV receptor ACE2 for entry and the serine protease TMPRSS2 for S protein priming. A TMPRSS2 inhibitor approved for clinical use blocked entry and might constitute a treatment option. Finally, we show that the sera from convalescent SARS patients cross-neutralized SARS-2-S-driven entry. Our results reveal important commonalities between SARS-CoV-2 and SARS-CoV infection and identify a potential target for antiviral intervention.
Keywords: SARS-CoV-2, COVID-19, ACE2, TMPRSS2, spike, entry, neutralization, coronavirus, priming
Graphical Abstract
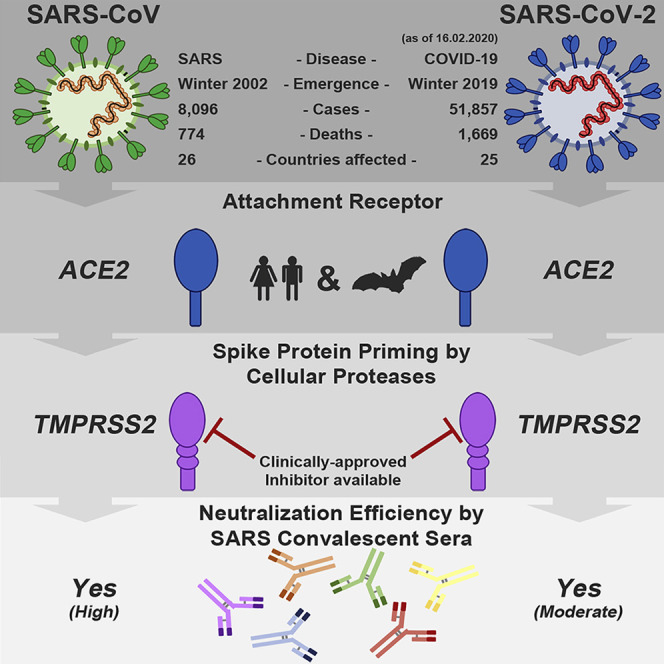
Highlights
-
•
SARS-CoV-2 uses the SARS-CoV receptor ACE2 for host cell entry
-
•
The spike protein of SARS-CoV-2 is primed by TMPRSS2
-
•
Antibodies against SARS-CoV spike may offer some protection against SARS-CoV-2
The emerging SARS-coronavirus 2 (SARS-CoV-2) threatens public health. Hoffmann and coworkers show that SARS-CoV-2 infection depends on the host cell factors ACE2 and TMPRSS2 and can be blocked by a clinically proven protease inhibitor. These findings might help to establish options for prevention and treatment.
Introduction
Several members of the family Coronaviridae constantly circulate in the human population and usually cause mild respiratory disease (Corman et al., 2019). In contrast, the severe acute respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV) are transmitted from animals to humans and cause severe respiratory diseases in afflicted individuals, SARS and MERS, respectively (Fehr et al., 2017). SARS emerged in 2002 in Guangdong province, China, and its subsequent global spread was associated with 8,096 cases and 774 deaths (de Wit et al., 2016, WHO, 2004). Chinese horseshoe bats serve as natural reservoir hosts for SARS-CoV (Lau et al., 2005, Li et al., 2005a). Human transmission was facilitated by intermediate hosts like civet cats and raccoon dogs, which are frequently sold as food sources in Chinese wet markets (Guan et al., 2003). At present, no specific antivirals or approved vaccines are available to combat SARS, and the SARS pandemic in 2002 and 2003 was finally stopped by conventional control measures, including travel restrictions and patient isolation.
In December 2019, a new infectious respiratory disease emerged in Wuhan, Hubei province, China (Huang et al., 2020, Wang et al., 2020, Zhu et al., 2020). An initial cluster of infections was linked to Huanan seafood market, potentially due to animal contact. Subsequently, human-to-human transmission occurred (Chan et al., 2020) and the disease, now termed coronavirus disease 19 (COVID-19) rapidly spread within China. A novel coronavirus, SARS-coronavirus 2 (SARS-CoV-2), which is closely related to SARS-CoV, was detected in patients and is believed to be the etiologic agent of the new lung disease (Zhu et al., 2020). On February 12, 2020, a total of 44,730 laboratory-confirmed infections were reported in China, including 8,204 severe cases and 1,114 deaths (WHO, 2020). Infections were also detected in 24 countries outside China and were associated with international travel. At present, it is unknown whether the sequence similarities between SARS-CoV-2 and SARS-CoV translate into similar biological properties, including pandemic potential (Munster et al., 2020).
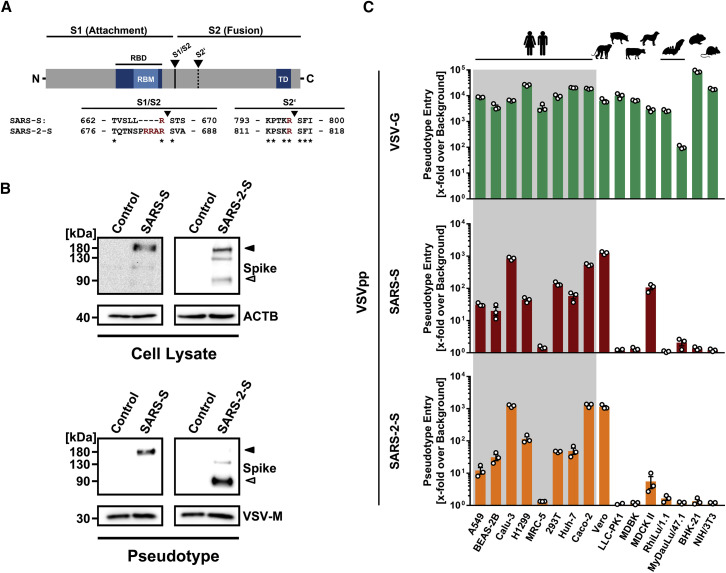
The spike (S) protein of coronaviruses facilitates viral entry into target cells. Entry depends on binding of the surface unit, S1, of the S protein to a cellular receptor, which facilitates viral attachment to the surface of target cells. In addition, entry requires S protein priming by cellular proteases, which entails S protein cleavage at the S1/S2 and the S2’ site and allows fusion of viral and cellular membranes, a process driven by the S2 subunit (Figure 1 A). SARS-S engages angiotensin-converting enzyme 2 (ACE2) as the entry receptor (Li et al., 2003) and employs the cellular serine protease TMPRSS2 for S protein priming (Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011). The SARS-S/ACE2 interface has been elucidated at the atomic level, and the efficiency of ACE2 usage was found to be a key determinant of SARS-CoV transmissibility (Li et al., 2005a, Li et al., 2005b). SARS-S und SARS-2-S share ∼76% amino acid identity. However, it is unknown whether SARS-2-S like SARS-S employs ACE2 and TMPRSS2 for host cell entry.
Figure 1.
SARS-2-S and SARS-S Facilitate Entry into a Similar Panel of Mammalian Cell Lines
(A) Schematic illustration of SARS-S including functional domains (RBD, receptor binding domain; RBM, receptor binding motif; TD, transmembrane domain) and proteolytic cleavage sites (S1/S2, S2′). Amino acid sequences around the two protease recognition sites (red) are indicated for SARS-S and SARS-2-S (asterisks indicate conserved residues). Arrow heads indicate the cleavage site.
(B) Analysis of SARS-2-S expression (upper panel) and pseudotype incorporation (lower panel) by western blot using an antibody directed against the C-terminal hemagglutinin (HA) tag added to the viral S proteins analyzed. Shown are representative blots from three experiments. β-Actin (cell lysates) and VSV-M (particles) served as loading controls (M, matrix protein). Black arrow heads indicate bands corresponding to uncleaved S proteins (S0) whereas gray arrow heads indicate bands corresponding to the S2 subunit.
(C) Cell lines of human and animal origin were inoculated with pseudotyped VSV harboring VSV-G, SARS-S, or SARS-2-S. At 16 h postinoculation, pseudotype entry was analyzed by determining luciferase activity in cell lysates. Signals obtained for particles bearing no envelope protein were used for normalization. The average of three independent experiments is shown. Error bars indicate SEM. Unprocessed data from a single experiment are presented in Figure S1.
Results
Evidence for Efficient Proteolytic Processing of SARS-2-S
The goal of our study was to obtain insights into how SARS-2-S facilitates viral entry into target cells and how this process can be blocked. For this, we first asked whether SARS-2-S is robustly expressed in a human cell line, 293T, commonly used for experimentation because of its high transfectability. Moreover, we analyzed whether there is evidence for proteolytic processing of the S protein because certain coronavirus S proteins are cleaved by host cell proteases at the S1/S2 cleavage site in infected cells (Figure 1A). Immunoblot analysis of 293T cells expressing SARS-2-S protein with a C-terminal antigenic tag revealed a band with a molecular weight expected for unprocessed S protein (S0) (Figure 1B). A band with a size expected for the S2 subunit of the S protein was also observed in cells and, more prominently, in vesicular stomatitis virus (VSV) particles bearing SARS-2-S (Figure 1B). In contrast, an S2 signal was largely absent in cells and particles expressing SARS-S (Figure 1B), as previously documented (Glowacka et al., 2011, Hofmann et al., 2004b). These results suggest efficient proteolytic processing of SARS-2-S in human cells, in keeping with the presence of several arginine residues at the S1/S2 cleavage site of SARS-2-S but not SARS-S (Figure 1A). In contrast, the S2′ cleavage site of SARS-2-S was similar to that of SARS-S.
SARS-2-S and SARS-S Mediate Entry into a Similar Spectrum of Cell Lines
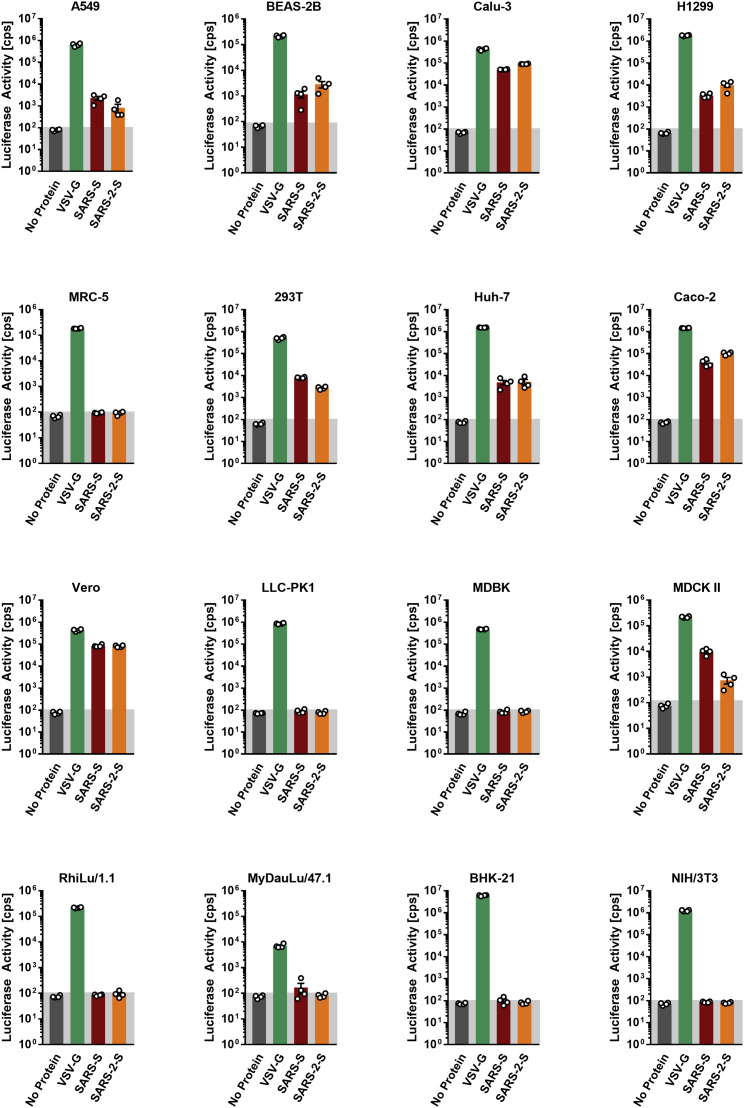
Replication-defective VSV particles bearing coronavirus S proteins faithfully reflect key aspects of coronavirus host cell entry (Kleine-Weber et al., 2019). We employed VSV pseudotypes bearing SARS-2-S to study cell entry of SARS-CoV-2. Both SARS-2-S and SARS-S were robustly incorporated into VSV particles (Figure 1B), allowing a meaningful side-by-side comparison; although, formally, comparable particle incorporation of the S1 subunit remains to be demonstrated. We first asked which cell lines were susceptible to SARS-2-S-driven entry, using a panel of well-characterized cell lines of human and animal origin, respectively. All cell lines were readily susceptible to entry driven by the glycoprotein of the pantropic VSV (VSV-G) (Figure 1C; Figure S1 ), as expected. Most human cell lines and the animal cell lines Vero and MDCKII were also susceptible to entry driven by SARS-S (Figure 1C). Moreover, SARS-2-S facilitated entry into an identical spectrum of cell lines as SARS-S (Figure 1C), suggesting similarities in choice of entry receptors.
Figure S1.
Representative Experiment Included in the Average, Related to Figure 1C
The indicated cells lines were inoculated with pseudoparticles harboring the indicated viral glycoprotein or harboring no glycoprotein (no protein) and luciferase activities in cell lysates were determined at 16 h posttransduction. The experiment was performed with quadruplicate samples, the average ± SD is shown.
SARS-CoV-2 Employs the SARS-CoV Receptor for Host Cell Entry
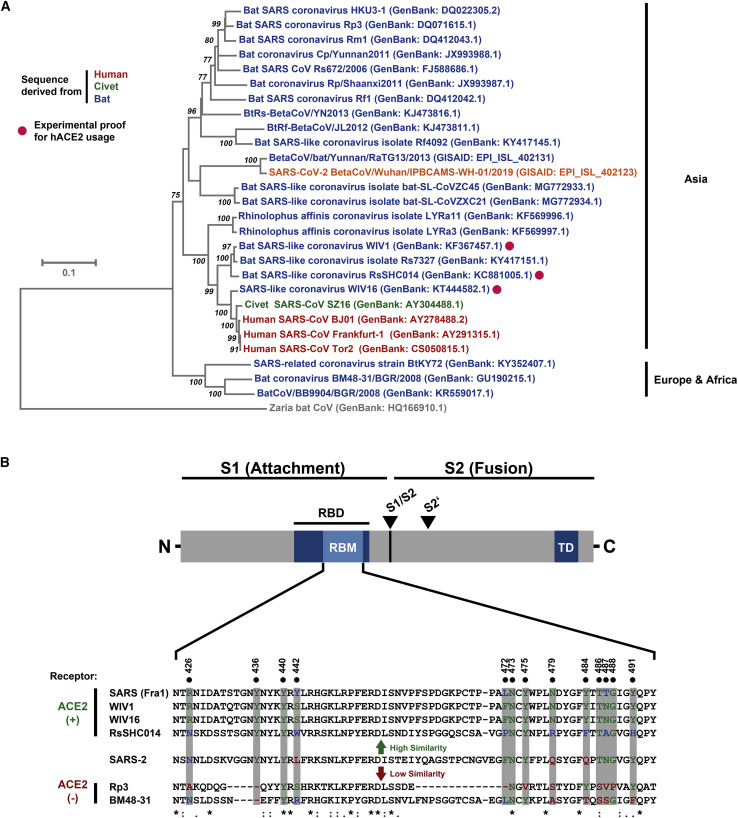
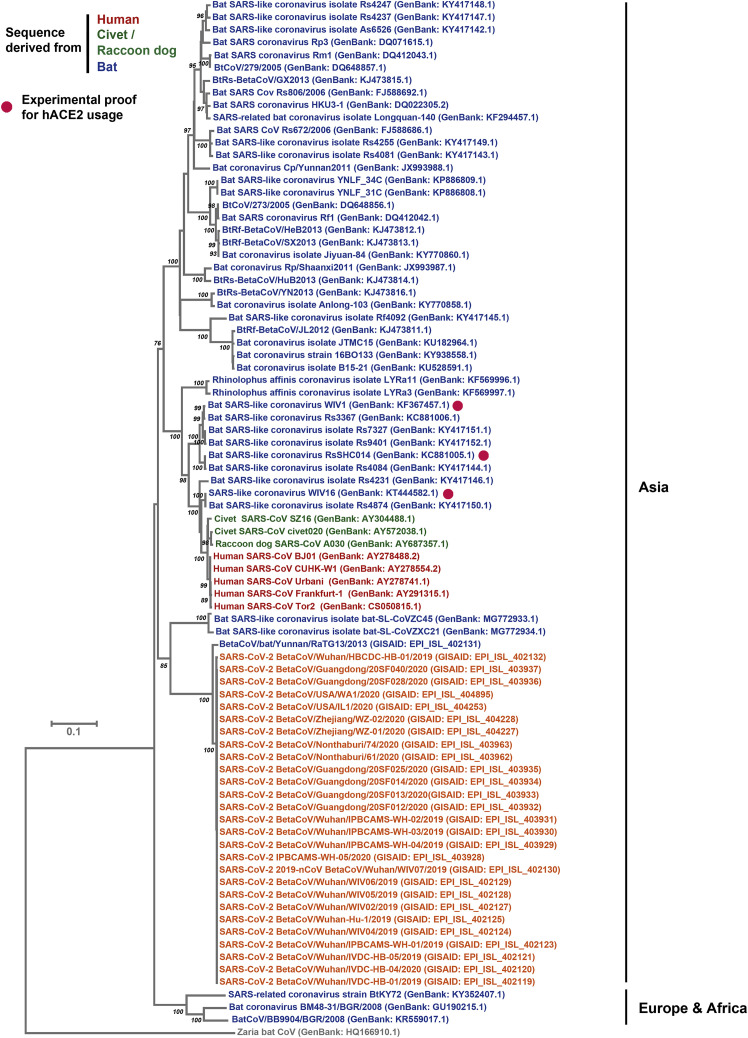
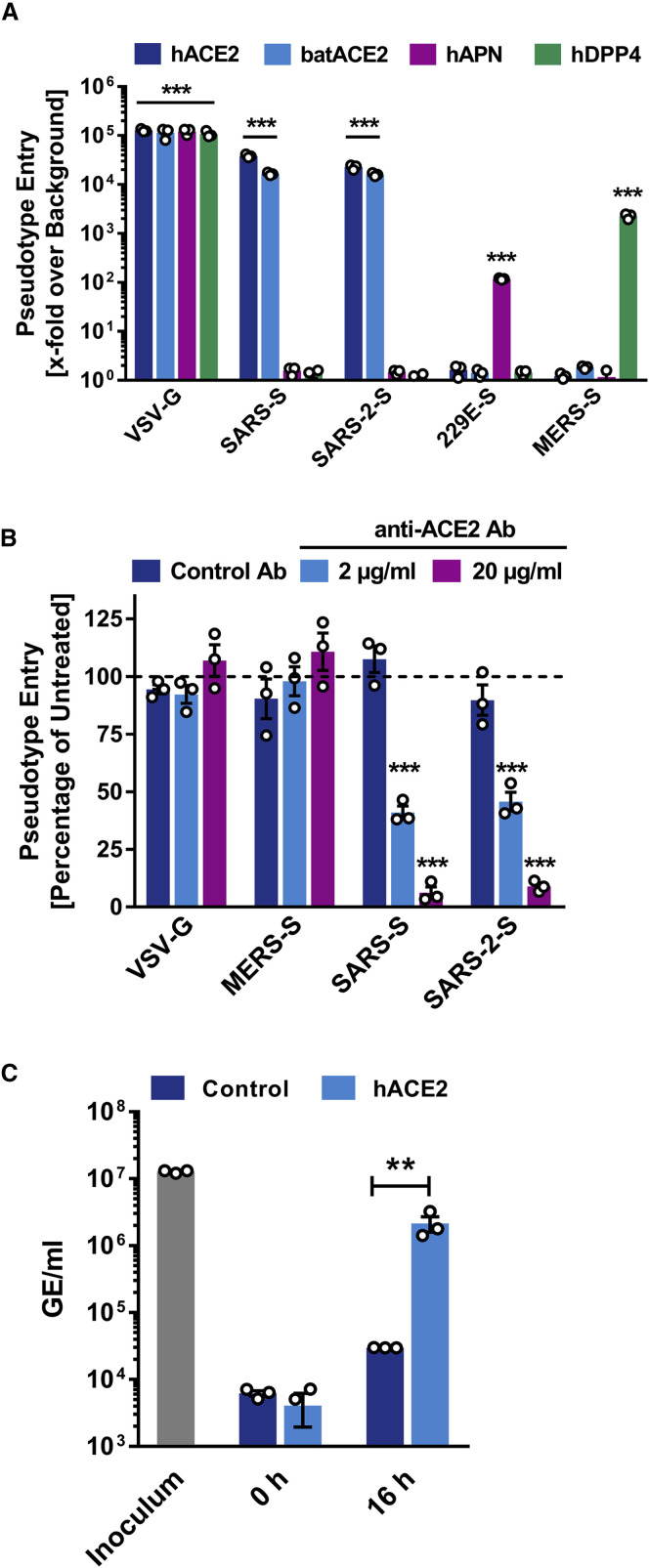
In order to elucidate why SARS-S and SARS-2-S mediated entry into the same cell lines, we next determined whether SARS-2-S harbors amino acid residues required for interaction with the SARS-S entry receptor ACE2. Sequence analysis revealed that SARS-CoV-2 clusters with SARS-CoV-related viruses from bats (SARSr-CoV), of which some but not all can use ACE2 for host cell entry (Figure 2 A; Figure S2 ). Analysis of the receptor binding motif (RBM), a portion of the receptor binding domain (RBD) that makes contact with ACE2 (Li et al., 2005a), revealed that most amino acid residues essential for ACE2 binding by SARS-S were conserved in SARS-2-S (Figure 2B). In contrast, most of these residues were absent from S proteins of SARSr-CoV previously found not to use ACE2 for entry (Figure 2B) (Ge et al., 2013, Hoffmann et al., 2013, Menachery et al., 2020). In agreement with these findings, directed expression of human and bat (Rhinolophus alcyone) ACE2 but not human DPP4, the entry receptor used by MERS-CoV (Raj et al., 2013), or human APN, the entry receptor used by HCoV-229E (Yeager et al., 1992), allowed SARS-2-S- and SARS-S-driven entry into otherwise non-susceptible BHK-21 cells (Figure 3 A). Moreover, antiserum raised against human ACE2 blocked SARS-S- and SARS-2-S- but not VSV-G- or MERS-S-driven entry (Figure 3B). Finally, authentic SARS-CoV-2 infected BHK-21 cells transfected to express ACE2 cells but not parental BHK-21 cells with high efficiency (Figure 3C), indicating that SARS-2-S, like SARS-S, uses ACE2 for cellular entry.
Figure 2.
SARS-2-S Harbors Amino Acid Residues Critical for ACE2 Binding
(A) The S protein of SARS-CoV-2 clusters phylogenetically with S proteins of known bat-associated betacoronaviruses (see Figure S2 for more details).
(B) Alignment of the receptor binding motif of SARS-S with corresponding sequences of bat-associated betacoronavirus S proteins, which are able or unable to use ACE2 as cellular receptor, reveals that SARS-CoV-2 possesses crucial amino acid residues for ACE2 binding.
Figure S2.
Extended Version of the Phylogenetic Tree, Related to Figure 2B
Figure 3.
SARS-2-S Utilizes ACE2 as Cellular Receptor
(A) BHK-21 cells transiently expressing ACE2 of human or bat origin, human APN, or human DPP4 were inoculated with pseudotyped VSV harboring VSV-G, SARS-S, SARS-2-S, MERS-S, or 229E-S. At 16 h postinoculation, pseudotype entry was analyzed (normalization against particles without viral envelope protein).
(B) Untreated Vero cells as well as Vero cells pre-incubated with 2 or 20 μg/mL of anti-ACE2 antibody or unrelated control antibody (anti-DC-SIGN, 20 μg/mL) were inoculated with pseudotyped VSV harboring VSV-G, SARS-S, SARS-2-S, or MERS-S. At 16 h postinoculation, pseudotype entry was analyzed (normalization against untreated cells).
(C) BHK-21 cells transfected with ACE2-encoding plasmid or control transfected with DsRed-encoding plasmid were infected with SARS-CoV-2 and washed, and genome equivalents in culture supernatants were determined by quantitative RT-PCR.
The average of three independent experiments conducted with triplicate samples is shown in (A–C). Error bars indicate SEM. Statistical significance was tested by two-way ANOVA with Dunnett posttest. Cells transfected with empty vector served as reference in (A) whereas cells that were not treated with antibody served as reference in (B).
The Cellular Serine Protease TMPRSS2 Primes SARS-2-S for Entry, and a Serine Protease Inhibitor Blocks SARS-CoV-2 Infection of Lung Cells
We next investigated protease dependence of SARS-CoV-2 entry. SARS-CoV can use the endosomal cysteine proteases cathepsin B and L (CatB/L) (Simmons et al., 2005) and the serine protease TMPRSS2 (Glowacka et al., 2011, Matsuyama et al., 2010, Shulla et al., 2011) for S protein priming in cell lines, and inhibition of both proteases is required for robust blockade of viral entry (Kawase et al., 2012). However, only TMPRSS2 activity is essential for viral spread and pathogenesis in the infected host whereas CatB/L activity is dispensable (Iwata-Yoshikawa et al., 2019, Shirato et al., 2016, Shirato et al., 2018, Zhou et al., 2015).
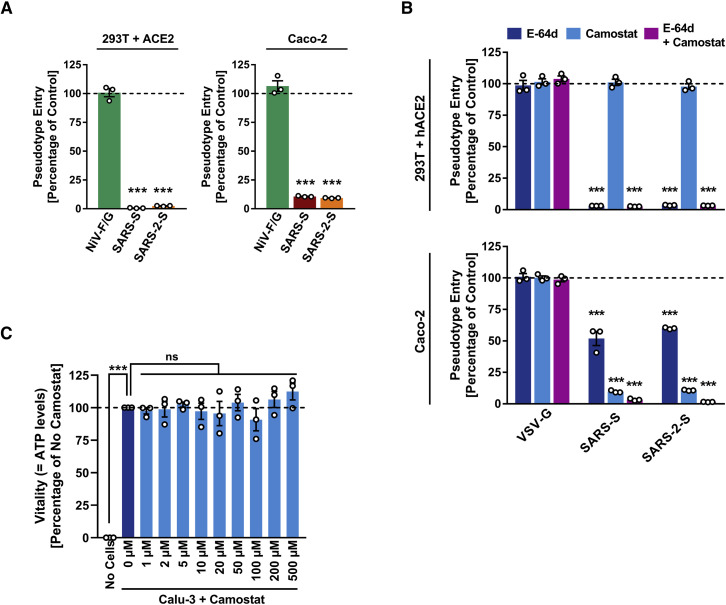
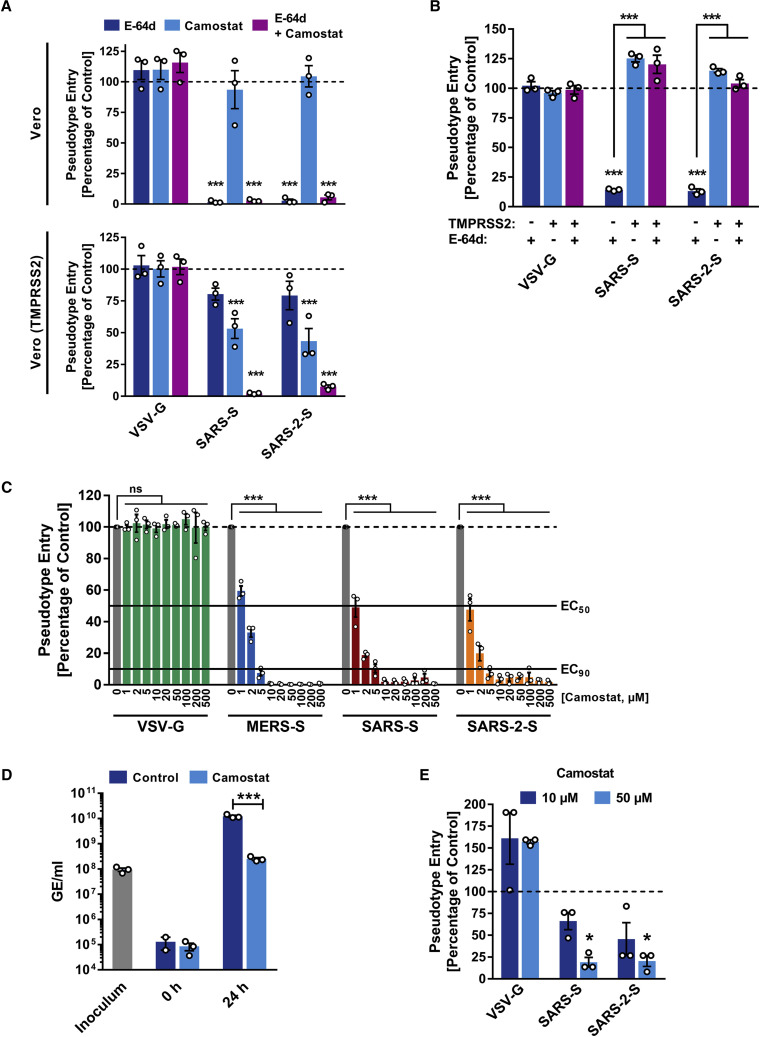
In order to determine whether SARS-CoV-2 can use CatB/L for cell entry, we initially employed ammonium chloride, which elevates endosomal pH and thereby blocks CatB/L activity. 293T cells (TMPRSS2−, transfected to express ACE2 for robust S protein-driven entry) and Caco-2 cells (TMPRSS2+) were used as targets. Ammonium chloride blocked VSV-G-dependent entry into both cell lines whereas entry driven by Nipah virus F and G proteins was not affected (Figure S3 A; data not shown), consistent with Nipah virus but not VSV being able to fuse directly with the plasma membrane (Bossart et al., 2002). Ammonium chloride treatment strongly inhibited SARS-2-S- and SARS-S-driven entry into TMPRSS2− 293T cells (Figure S3 A), suggesting CatB/L dependence. Inhibition of entry into TMPRSS2+ Caco-2 cells was less efficient compared to 293T cells (Figure S3 A), which would be compatible with SARS-2-S priming by TMPRSS2 in Caco-2 cells. Indeed, the clinically proven serine protease inhibitor camostat mesylate, which is active against TMPRSS2 (Kawase et al., 2012), partially blocked SARS-2-S-driven entry into Caco-2 (Figure S3 B) and Vero-TMPRSS2 cells (Figure 4 A). Full inhibition was attained when camostat mesylate and E-64d, an inhibitor of CatB/L, were added (Figure 4A; Figure S3B), indicating that SARS-2-S can use both CatB/L as well as TMPRSS2 for priming in these cell lines. In contrast, camostat mesylate did not interfere with SARS-2-S-driven entry into the TMPRSS2− cell lines 293T (Figure S3B) and Vero (Figure 4A), which was efficiently blocked by E-64d and therefore is CatB/L dependent. Moreover, directed expression of TMPRSS2 rescued SARS-2-S-driven entry from inhibition by E-64d (Figure 4B), confirming that SARS-2-S can employ TMPRSS2 for S protein priming.
Figure S3.
Protease Requirement for SARS-2-S-Driven Entry and Absence of Unwanted Cytotoxicity of Camostat Mesylate, Related to Figure 4
(A and B) Importance of endosomal low pH (A) and activity of CatB/L or TMPRSS2 (B) for host cell entry of SARS-CoV-2 was evaluated by adding inhibitors to target cells prior to transduction. Ammonium chloride (A) blocks endosomal acidification while E-64d and camostat mesylate (B) block the activity of CatB/L and TMPRSS2, respectively. Entry into cells not treated with inhibitor was set as 100%.
(C) Absence of cytotoxic effects of camostat mesylate. Calu-3 cells were treated with camostat mesylate identically as for infection experiments and cell viability was measured using a commercially available assay (CellTiter-Glo, Promega).
Figure 4.
SARS-2-S Employs TMPRSS2 for S Protein Priming in Human Lung Cells
(A) Importance of activity of CatB/L or TMPRSS2 for host cell entry of SARS-CoV-2 was evaluated by adding inhibitors to target cells prior to transduction. E-64d and camostat mesylate block the activity of CatB/L and TMPRSS2, respectively (additional data for 293T cells transiently expressing ACE2 and Caco-2 cells are shown in Figure S3).
(B) To analyze whether TMPRSS2 can rescue SARS-2-S-driven entry into cells that have low CatB/L activity, 293T cells transiently expressing ACE2 alone or in combination with TMPRSS2 were incubated with CatB/L inhibitor E-64d or DMSO as control and inoculated with pseudotypes bearing the indicated viral surface proteins.
(C) Calu-3 cells were pre-incubated with the indicated concentrations of camostat mesylate and subsequently inoculated with pseudoparticles harboring the indicated viral glycoproteins.
(D) Calu-3 cells were pre-incubated with camostat mesylate and infected with SARS-CoV-2. Subsequently, the cells were washed and genome equivalents in culture supernatants were determined by quantitative RT-PCR.
(E) In order to investigate whether serine protease activity is required for SARS-2-S-driven entry into human lung cells, primary human airway epithelial cells were incubated with camostat mesylate prior to transduction.
The average of three independent experiments conducted with triplicate or quadruplicate samples is shown in (A–E). Error bars indicate SEM. Statistical significance was tested by two-way ANOVA with Dunnett posttest. Cells that did not receive inhibitor served as reference in (A), (C), (D), and (E) whereas cells transfected with empty vector and not treated with inhibitor served as reference in (B).
We next analyzed whether TMPRSS2 usage is required for SARS-CoV-2 infection of lung cells. Indeed, camostat mesylate significantly reduced MERS-S-, SARS-S-, and SARS-2-S- but not VSV-G-driven entry into the lung cell line Calu-3 (Figure 4C) and exerted no unwanted cytotoxic effects (Figure S3 C). Similarly, camostat mesylate treatment significantly reduced Calu-3 infection with authentic SARS-CoV-2 (Figure 4D). Finally, camostat mesylate treatment inhibited SARS-S- and SARS-2-S- but not VSV-G-driven entry into primary human lung cells (Figure 4E). Collectively, SARS-CoV-2 can use TMPRSS2 for S protein priming and camostat mesylate, an inhibitor of TMPRSS2, blocks SARS-CoV-2 infection of lung cells.
Evidence that Antibodies Raised against SARS-CoV Will Cross-Neutralize SARS-CoV-2
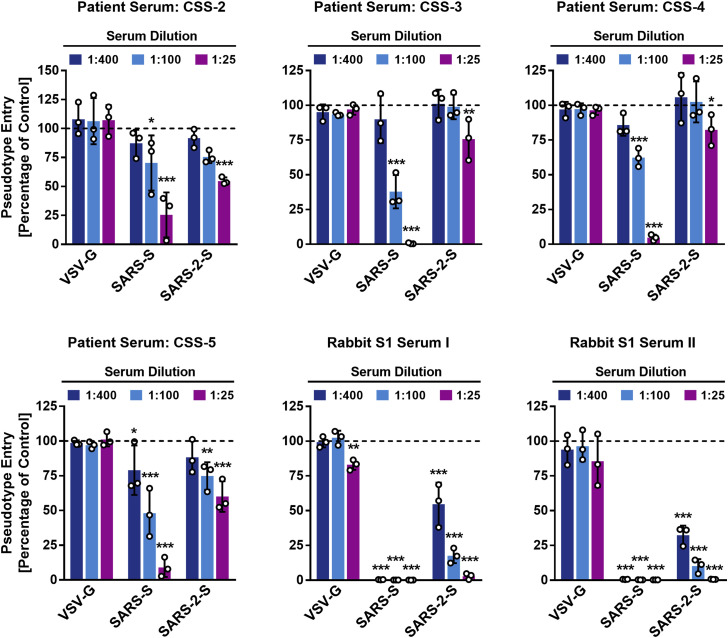
Convalescent SARS patients exhibit a neutralizing antibody response directed against the viral S protein (Liu et al., 2006). We investigated whether such antibodies block SARS-2-S-driven entry. Four sera obtained from three convalescent SARS patients inhibited SARS-S- but not VSV-G-driven entry in a concentration-dependent manner (Figure 5 ). In addition, these sera also reduced SARS-2-S-driven entry, although with lower efficiency compared to SARS-S (Figure 5). Similarly, rabbit sera raised against the S1 subunit of SARS-S reduced both SARS-S- and SARS-2-S-driven entry with high efficiency, and again inhibition of SARS-S-driven entry was more efficient. Thus, antibody responses raised against SARS-S during infection or vaccination might offer some level of protection against SARS-CoV-2 infection.
Figure 5.
Sera from Convalescent SARS Patients Cross-Neutralize SARS-2-S-Driven Entry
Pseudotypes harboring the indicated viral surface proteins were incubated with different dilutions of sera from three convalescent SARS patients or sera from rabbits immunized with the S1 subunit of SARS-S and subsequently inoculated onto Vero cells in order to evaluate cross-neutralization potential. The average of three independent experiments performed with triplicate samples is shown. Error bars indicate SEM. Statistical significance was tested by two-way ANOVA with Dunnett posttest.
Discussion
The present study provides evidence that host cell entry of SARS-CoV-2 depends on the SARS-CoV receptor ACE2 and can be blocked by a clinically proven inhibitor of the cellular serine protease TMPRSS2, which is employed by SARS-CoV-2 for S protein priming. Moreover, it suggests that antibody responses raised against SARS-CoV could at least partially protect against SARS-CoV-2 infection. These results have important implications for our understanding of SARS-CoV-2 transmissibility and pathogenesis and reveal a target for therapeutic intervention.
The finding that SARS-2-S exploits ACE2 for entry, which was also reported by Zhou and colleagues (Zhou et al., 2020) while the present manuscript was in revision, suggests that the virus might target a similar spectrum of cells as SARS-CoV. In the lung, SARS-CoV infects mainly pneumocytes and macrophages (Shieh et al., 2005). However, ACE2 expression is not limited to the lung, and extrapulmonary spread of SARS-CoV in ACE2+ tissues was observed (Ding et al., 2004, Gu et al., 2005, Hamming et al., 2004). The same can be expected for SARS-CoV-2, although affinity of SARS-S and SARS-2-S for ACE2 remains to be compared. It has been suggested that the modest ACE2 expression in the upper respiratory tract (Bertram et al., 2012, Hamming et al., 2004) might limit SARS-CoV transmissibility. In light of the potentially increased transmissibility of SARS-CoV-2 relative to SARS-CoV, one may speculate that the new virus might exploit cellular attachment-promoting factors with higher efficiency than SARS-CoV to ensure robust infection of ACE2+ cells in the upper respiratory tract. This could comprise binding to cellular glycans, a function ascribed to the S1 domain of certain coronaviruses (Li et al., 2017, Park et al., 2019). Finally, it should be noted that ACE2 expression protects from lung injury and is downregulated by SARS-S (Haga et al., 2008, Imai et al., 2005, Kuba et al., 2005), which might promote SARS. It will thus be interesting to determine whether SARS-CoV-2 also interferes with ACE2 expression.
Priming of coronavirus S proteins by host cell proteases is essential for viral entry into cells and encompasses S protein cleavage at the S1/S2 and the S2′ sites. The S1/S2 cleavage site of SARS-2-S harbors several arginine residues (multibasic), which indicates high cleavability. Indeed, SARS-2-S was efficiently cleaved in cells, and cleaved S protein was incorporated into VSV particles. Notably, the cleavage site sequence can determine the zoonotic potential of coronaviruses (Menachery et al., 2020, Yang et al., 2014, Yang et al., 2015), and a multibasic cleavage site was not present in RaTG13, the coronavirus most closely related to SARS-CoV-2. It will thus be interesting to determine whether the presence of a multibasic cleavage site is required for SARS-CoV-2 entry into human cells and how this cleavage site was acquired.
The S proteins of SARS-CoV can use the endosomal cysteine proteases CatB/L for S protein priming in TMPRSS2− cells (Simmons et al., 2005). However, S protein priming by TMPRSS2 but not CatB/L is essential for viral entry into primary target cells and for viral spread in the infected host (Iwata-Yoshikawa et al., 2019, Kawase et al., 2012, Zhou et al., 2015). The present study indicates that SARS-CoV-2 spread also depends on TMPRSS2 activity, although we note that SARS-CoV-2 infection of Calu-3 cells was inhibited but not abrogated by camostat mesylate, likely reflecting residual S protein priming by CatB/L. One can speculate that furin-mediated precleavage at the S1/S2 site in infected cells might promote subsequent TMPRSS2-dependent entry into target cells, as reported for MERS-CoV (Kleine-Weber et al., 2018, Park et al., 2016). Collectively, our present findings and previous work highlight TMPRSS2 as a host cell factor that is critical for spread of several clinically relevant viruses, including influenza A viruses and coronaviruses (Gierer et al., 2013, Glowacka et al., 2011, Iwata-Yoshikawa et al., 2019, Kawase et al., 2012, Matsuyama et al., 2010, Shulla et al., 2011, Zhou et al., 2015). In contrast, TMPRSS2 is dispensable for development and homeostasis (Kim et al., 2006) and thus constitutes an attractive drug target. In this context, it is noteworthy that the serine protease inhibitor camostat mesylate, which blocks TMPRSS2 activity (Kawase et al., 2012, Zhou et al., 2015), has been approved in Japan for human use, but for an unrelated indication. This compound or related ones with potentially increased antiviral activity (Yamamoto et al., 2016) could thus be considered for off-label treatment of SARS-CoV-2-infected patients.
Convalescent SARS patients exhibit a neutralizing antibody response that can be detected even 24 months after infection (Liu et al., 2006) and that is largely directed against the S protein. Moreover, experimental SARS vaccines, including recombinant S protein (He et al., 2006) and inactivated virus (Lin et al., 2007), induce neutralizing antibody responses. Although confirmation with infectious virus is pending, our results indicate that neutralizing antibody responses raised against SARS-S could offer some protection against SARS-CoV-2 infection, which may have implications for outbreak control.
In sum, this study provided key insights into the first step of SARS-CoV-2 infection, viral entry into cells, and defined potential targets for antiviral intervention.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-HA antibody produced in mouse | Sigma-Aldrich | Cat.#: H3663; RRID: AB_262051 |
| Monoclonal anti-β-actin antibody produced in mouse | Sigma-Aldrich | Cat.#: A5441; RRID: AB_476744 |
| Monoclonal anti-VSV-M (23H12) antibody | KeraFast | Cat.#: EB0011; RRID: AB_2734773 |
| Polyclonal anti-ACE2 antibody | R&D Systems | Cat.#: AF933; RRID: AB_355722 |
| Polyclonal anti-DC-SIGN antibody | Santa Cruz | Cat.#: sc-11038; RRID:AB_639038 |
| Monoclonal anti-mouse, peroxidase-coupled | Dianova | Cat.#: 115-035-003; RRID:AB_10015289 |
| Anti-VSV-G antibody (I1, produced from CRL-2700 mouse hybridoma cells) | ATCC | Cat.# CRL-2700; RRID:CVCL_G654 |
| Bacterial and Virus Strains | ||
| VSV∗ΔG-FLuc | (Berger Rentsch and Zimmer, 2011) | N/A |
| SARS-CoV-2 isolate Munich 929 | Laboratory of Christian Drosten | N/A |
| One Shot™ OmniMAX™ 2 T1R Chemically Competent E. coli | ThermoFisher Scientific | Cat.#: C854003 |
| Biological Samples | ||
| Patient serum, CSS-2 | Laboratory of Christian Drosten | N/A |
| Patient serum, CSS-3 | Laboratory of Andreas Nitsche | N/A |
| Patient serum, CSS-4 | Laboratory of Andreas Nitsche | N/A |
| Patient serum, CSS-5 | Laboratory of Andreas Nitsche | N/A |
| Rabbit serum, anti-SARS-S1 rabbit I | Laboratory of Stefan Pöhlmann | N/A |
| Rabbit serum, anti-SARS-S1 rabbit II | Laboratory of Stefan Pöhlmann | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Camostat mesylate | Sigma-Aldrich | SML0057 |
| E-64d | Sigma-Aldrich | E8640 |
| Ammonium chloride | Carl Roth | Cat.#: 5050.2 |
| Critical Commercial Assays | ||
| Beetle-Juice Kit | PJK | Cat.#: 102511 |
| CellTiter-Glo® Luminescent Cell Viability Assay | Promega | Cat.#: G7570 |
| Experimental Models: Cell Lines | ||
| A549 | Laboratory of Georg Herrler | ATCC Cat# CRM-CCL-185; RRID:CVCL_0023 |
| BEAS-2B | Laboratory of Stefan Pöhlmann | ATCC Cat# CRL-9609; RRID:CVCL_0168 |
| Calu-3 | Laboratory of Stephan Ludwig | ATCC Cat# HTB-55; RRID:CVCL_0609 |
| NCI-H1299 | Laboratory of Stefan Pöhlmann | ATCC Cat# CRL-5803; RRID:CVCL_0060 |
| Huh-7 | Laboratory of Thomas Pietschmann | JCRB Cat# JCRB0403; RRID:CVCL_0336 |
| Caco-2 | Laboratory of Stefan Pöhlmann | ATCC Cat# HTB-37; RRID:CVCL_0025 |
| Vero | Laboratory of Andrea Maisner | ATCC Cat# CRL-1586; RRID:CVCL_0574 |
| Vero-TMPRSS2 | This paper | N/A |
| LLC-PK1 | Laboratory of Georg Herrler | ATCC Cat# CRL-1392; RRID:CVCL_0391 |
| MDBK | Laboratory of Georg Herrler | ATCC Cat# CCL-22; RRID:CVCL_0421 |
| MDCKII | Laboratory of Georg Herrler | ATCC Cat# CRL-2936; RRID:CVCL_B034 |
| RhiLu/1.1 | Laboratory of Christian Drosten, Laboratory of Marcel A. Müller | N/A; RRID: CVCL_RX22 |
| MyDauLu/47.1 | Laboratory of Christian Drosten, Laboratory of Marcel A. Müller | N/A; RRID: CVCL_RX18 |
| BHK-21 | Laboratory of Georg Herrler | ATCC Cat# CCL-10; RRID:CVCL_1915 |
| NIH/3T3 | Laboratory of Stefan Pöhlmann | ATCC Cat# CRL-1658; RRID:CVCL_0594 |
| HAE | HTCR Foundation (Human Tissue and Cell Research) | N/A |
| 293T | DSMZ | Cat.#: ACC-635; RRID: CVCL_0063 |
| Oligonucleotides | ||
| SARS-2-S (BamHI) F AAGGCCGGATCCGCCACCATGTTTCT GCTGACCACCAAGC |
Sigma-Aldrich | N/A |
| SARS-2-S (XbaI) R AAGGCCTCTAGATTAGGTGTAGTGCAG TTTCACG |
Sigma-Aldrich | N/A |
| SARS-2-S-HA (XbaI) R AAGGCCTCTAGATTACGCATAATCC GGCACATCATACGGATAGGTGTAGTGCAGTTTCACG |
Sigma-Aldrich | N/A |
| WH-Ssyn 651F CAAGATCTACAGCAAGCACACC | Sigma-Aldrich | N/A |
| WH-Ssyn 1380F GTCGGCGGCAACTACAATTAC | Sigma-Aldrich | N/A |
| WH-Ssyn 1992F CTGTCTGATCGGAGCCGAGCAC | Sigma-Aldrich | N/A |
| WH-Ssyn 2648F TGAGATGATCGCCCAGTACAC | Sigma-Aldrich | N/A |
| WH-Ssyn 3286F GCCATCTGCCACGACGGCAAAG | Sigma-Aldrich | N/A |
| Recombinant DNA | ||
| Synthetic, codon-optimized (humanized) SARS-2-S | ThermoFisher Scientific (GeneArt) | N/A |
| Plasmid: pCG1-SARS-S | (Hoffmann et al., 2013) | N/A |
| Plasmid:pCG1-SARS-S-HA | This paper | N/A |
| Plasmid: pCG1-SARS-2-S | This paper | N/A |
| Plasmid: pCG1-SARS-2-S-HA | This paper | N/A |
| Plasmid: pCAGGS-229E-S | (Hofmann et al., 2005) | N/A |
| Plasmid: pCAGGS-MERS-S | (Gierer et al., 2013) | N/A |
| Plasmid: pCAGGS-VSV-G | (Brinkmann et al., 2017) | N/A |
| Plasmid: pCAGGS-NiV-F | Laboratory of Andrea Maisner | N/A |
| Plasmid: pCAGGS-NiV-G | Laboratory of Andrea Maisner | N/A |
| Plasmid: pCG1-hACE2 | (Hoffmann et al., 2013) | N/A |
| Plasmid: pCG1-batACE2 | (Hoffmann et al., 2013) | N/A |
| Plasmid: pCG1-hAPN | (Hofmann et al., 2004a) | N/A |
| Plasmid: pQCXIP-DsRed-hDPP4 | (Kleine-Weber et al., 2018) | N/A |
| Plasmid: pQCXIBL-hTMPRSS2 | (Kleine-Weber et al., 2018) | N/A |
| Plasmid: pCG1 | Laboratory of Roberto Cattaneo | N/A |
| Plasmid: pCAGGS-DsRed | Laboratory of Stefan Pöhlmann | N/A |
| Plasmid: pCAGGS-eGFP | Laboratory of Stefan Pöhlmann | N/A |
| Software and Algorithms | ||
| Hidex Sense Microplate Reader Software | Hidex Deutschland Vertrieb GmbH | https://www.hidex.de/ |
| ChemoStar Imager Software (version v.0.3.23) | Intas Science Imaging Instruments GmbH | https://www.intas.de/ |
| MEGA 7.0.26 | Kumar et al., 2018 | https://www.megasoftware.net |
| Adobe Photoshop CS5 Extended (version 12.0 × 32) | Adobe | https://www.adobe.com/ |
| GraphPad Prism (version 8.3.0(538)) | GraphPad Software | https://www.graphpad.com/ |
| Microsoft Office Standard 2010 (version 14.0.7232.5000) | Microsoft Corporation | https://products.office.com/ |
Lead Contact and Materials Availability
Requests for material can be directed to Markus Hoffmann (mhoffmann@dpz.eu) and the lead contact, Stefan Pöhlmann (spoehlmann@dpz.eu). All materials and reagents will be made available upon installment of a material transfer agreement (MTA).
Experimantal Model and Subject Details
Cell cultures, primary cells, viral strains
All cell lines were incubated at 37°C and 5% CO2 in a humidified atmosphere. 293T (human, kidney), BHK-21 (Syrian hamster, kidney cells), Huh-7 (human, liver), LLC-PK1 (pig, kidney), MRC-5 (human, lung), MyDauLu/47.1 (Daubenton’s bat [Myotis daubentonii], lung), NIH/3T3 (Mouse, embryo), RhiLu/1.1 (Halcyon horseshoe bat [Rhinolophus alcyone], lung) and Vero (African green monkey, kidney) cells were incubated in Dulbecco’s’ modified Eagle medium (PAN-Biotech). Calu-3 (human, lung), Caco-2 (human, colon), MDBK (cattle, kidney) and MDCKII (Dog, kidney) cells were incubated in Minimum Essential Medium (ThermoFisher Scientific). A549 (human, lung), BEAS-2B (human, bronchus) and NCI-H1299 (human, lung) cells were incubated in DMEM/F-12 Medium with Nutrient Mix (ThermoFisher Scientific). Vero cells stably expressing human TMPRSS2 were generated by retroviral transduction and blasticidin-based selection. All media were supplemented with 10% fetal bovine serum (Biochrom), 100 U/mL of penicillin and 0.1 mg/mL of streptomycin (PAN-Biotech), 1x non-essential amino acid solution (10x stock, PAA) and 10 mM sodium pyruvate (ThermoFisher Scientific). For seeding and subcultivation, cells were first washed with phosphate buffered saline (PBS) and then incubated in the presence of trypsin/EDTA solution (PAN-Biotech) until cells detached. Transfection was carried out by calcium-phosphate precipitation. Lung tissue samples were obtained and experimental procedures were performed within the framework of the non-profit foundation HTCR, including the informed patient’s consent.
For preparation of human airway epithelial cells, bronchus tissue was derived from patients undergoing pulmonary resection and was provided by the Biobank of the Department of General, Visceral, and Transplant Surgery, Ludwig-Maximilians- University Munich. Primary human airway epithelial cells were subsequently isolated as described (Wu et al., 2016). In brief, tissue with a length of approximately 10 mm and a diameter of 8mm was collected and incubated for 24 h at 4°C with DMEM (GIBCO) containing 1 mg/mL protease type XIV and 10 μg/mL DNase I, 100 units/mL penicillin and 100 μg/mL streptomycin, 2.5 μg/mL amphotericin B, and 50 μg/mL gentamicin (GIBCO). The epithelial cells were then harvested from the mucosal surface using the scalpel and were resuspended in growth medium. After incubation at 37°C, 5% CO2 for 2 h to remove adherent fibroblast cells, non-adherent cells were seeded on a collagen I coated flask and maintained at 37°C, 5% CO2. The growth medium was refreshed every 2 days and consisted of a 1:1 mixture of DMEM (GIBCO) and Airway Epithelial Cell basal medium (Promocell, Heidelberg, Germany) supplemented with 52 μg/mL bovine pituitary extract, 15 ng/mL retinoic acid, 5μg/mL insulin, 0.5 μg/mL hydrocortisone, 0.5 μg/mL epinephrine, 10 μg/mL transferrin, 1 ng/mL human epidermal growth factor (Corning), 1.5 ng/mL bovine serum albumin, 100 units/mL penicillin and 100 μg/mL streptomycin, with or without 5 μM Rho-associated protein kinase inhibitor (Y-27632), as previously described (Wu et al., 2016). If not stated otherwise all materials were purchased from Sigma-Aldrich.
For infection experiments with SARS-CoV-2, the SARS-CoV-2 isolate Munich 929 was propagated in VeroE6 cells (passage 1) after primary isolation from patient material on Vero-TMPRSS2 cells (passage 0).
Method Details
Plasmids
Expression plasmids for vesicular stomatitis virus (VSV, serotype Indiana) glycoprotein (VSV-G), Nipah virus (NiV) fusion (F) and attachment glycoprotein (G), SARS-S (derived from the Frankfurt-1 isolate) with or without a C-terminal HA epitope tag, HCoV-229E-S, MERS-S, human and bat angiotensin converting enzyme 2 (ACE2), human aminopeptidase N (APN), human dipeptidyl-peptidase 4 (DPP4) and human TMPRSS2 have been described elsewhere (Bertram et al., 2010, Brinkmann et al., 2017, Gierer et al., 2013, Hoffmann et al., 2013, Hofmann et al., 2005, Kleine-Weber et al., 2019). For generation of the expression plasmids for SARS-2-S with or without a C-terminal HA epitope tag we PCR-amplified the coding sequence of a synthetic, codon-optimized (for human cells) SARS-2-S DNA (GeneArt Gene Synthesis, ThermoFisher Scientific) based on the publicly available protein sequence in the National Center for Biotechnology Information database (NCBI Reference Sequence: YP_009724390.1) and cloned in into the pCG1 expression vector via BamHI and XbaI restriction sites.
Pseudotyping of VSV and transduction experiments
For pseudotyping, VSV pseudotypes were generated according to a published protocol (Berger Rentsch and Zimmer, 2011). In brief, 293T transfected to express the viral surface glycoprotein under study were inoculated with a replication-deficient VSV vector that contains expression cassettes for eGFP (enhanced green fluorescent protein) and firefly luciferase instead of the VSV-G open reading frame, VSV∗ΔG-fLuc (kindly provided by Gert Zimmer, Institute of Virology and Immunology, Mittelhäusern/Switzerland). After an incubation period of 1 h at 37°C, the inoculum was removed and cells were washed with PBS before medium supplemented with anti-VSV-G antibody (I1, mouse hybridoma supernatant from CRL-2700; ATCC) was added in order to neutralize residual input virus (no antibody was added to cells expressing VSV-G). Pseudotyped particles were harvested 16 h postinoculation, clarified from cellular debris by centrifugation and used for experimentation.
For transduction, target cells were grown in 96-well plates until they reached 50%–75% confluency before they were inoculated with respective pseudotyped VSV. For experiments addressing receptor usage, cells were transfected with expression plasmids 24 h before transduction. In order to block ACE2 on the cell surface, cells were pretreated with 2 or 20 μg/mL anti-ACE2 antibody (R&D Systems, goat, AF933). As control, an unrelated anti-DC-SIGN antibody (Serotec, goat, 20 μg/mL) was used. For experiments involving ammonium chloride (final concentration 50 mM) and protease inhibitors (E-64d, 25 μM; camostat mesylate, 1-500 μM), target cells were treated with the respective chemical 2 h before transduction. For neutralization experiments, pseudotypes were pre-incubated for 30 min at 37°C with different serum dilutions. Transduction efficiency was quantified 16 h posttransduction by measuring the activity of firefly luciferase in cell lysates using a commercial substrate (Beetle-Juice, PJK) and a Hidex Sense plate luminometer (Hidex).
Quantification of cell viability
Cell viability following treatment of Calu-3 cells with camostat mesylate was analyzed using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega). In brief, Calu-3 cells grown to 50% confluency in 96-well plates were incubated for 24 h in the absence or presence of different concentrations (1-500 μM) of camostat mesylate. Next, the culture medium was aspirated and 100 μl of fresh culture medium was added before an identical volume of the assay substrate was added. Wells containing only culture medium served as a control to determine the assay background. After 2 min of incubation on a rocking platform and additional 10 min without movement, samples were transferred into white opaque-walled 96-well plates and luminescent signal were recorded using a Hidex Sense plate luminometer (Hidex).
Analysis of SARS-2-S expression and particle incorporation
To analyze S protein expression in cells, 293T cells were transfected with expression vectors for HA-tagged SARS-2-S or SARS-S or empty expression vector (negative control). The culture medium was replaced at 16 h posttransfection and the cells were incubated for an additional 24 h. Then, the culture medium was removed and cells were washed once with PBS before 2x SDS-sample buffer (0.03 M Tris-HCl, 10% glycerol, 2% SDS, 0.2% bromophenol blue, 1 mM EDTA) was added and cells were incubated for 10 min at room temperature. Next, the samples were heated for 15 min at 96°C and subjected to SDS-PAGE and immunoblotting.
For analysis of S protein incorporation into pseudotyped particles, 1 mL of the respective VSV pseudotypes were loaded onto a 20% (w/v) sucrose cushion (volume 50 μl) and subjected to high-speed centrifugation (25.000 g for 120 min at 4°C). Thereafter, 1 mL of supernatant was removed and the residual volume was mixed with 50 μl of 2x SDS-sample buffer, heated for 15 min at 96°C and subjected to SDS-PAGE and immunoblotting. After protein transfer, nitrocellulose membranes were blocked in 5% skim milk solution (5% skim milk dissolved in PBS containing 0.05% Tween-20, PBS-T) for 1 h at room temperature and then incubated over night at 4°C with the primary antibody (diluted in in skim milk solution)). Following three washing intervals of 10 min in PBS-T the membranes were incubated for 1 h at room temperature with the secondary antibody (diluted in in skim milk solution), before the membranes were washed and imaged using an in in house-prepared enhanced chemiluminescent solution (0.1 M Tris-HCl [pH 8.6], 250 μg/mL luminol, 1 mg/mL para-hydroxycoumaric acid, 0.3% H2O2) and the ChemoCam imaging system along with the ChemoStar Professional software (Intas Science Imaging Instruments GmbH). The following primary antibodies were used: Mouse anti-HA tag (Sigma-Aldrich, H3663, 1:2,500), mouse anti-β-actin (Sigma-Aldrich, A5441, 1:2,000), mouse anti-VSV matrix protein (Kerafast, EB0011, 1:2,500). As secondary antibody we used a peroxidase-coupled goat anti-mouse antibody (Dianova, 115-035-003, 1:10000).
Infection with authentic SARS-CoV-2
BHK-21 cells (1.6 x105 cells/mL) were transfected with ACE2 and DsRed as a negative control. After 24 h, cells were washed with PBS and infected with 8x107 genome equivalents (GE) per 24-well of SARS-CoV-2 isolate Munich 929 for 1 h at 37°C. Calu-3 cells (5 x105 cells/mL) were mock treated or treated with 100 μM camostat mesylate (Sigma Aldrich) 2 h prior to infection with SARS-CoV-2 isolate Munich 929 at a multiplicity of infection (MOI) of 0.001 for 1 h at 37°C. After infection, cells were washed three times with PBS before 500 μl of DMEM medium was added. At 16 or 24 h post infection, 50 μl culture supernatant was subjected to viral RNA extraction using a viral RNA kit (Macherey-Nagel) according to the manufacturer’s instructions. GE per ml were detected by real time RT-PCR using a previously reported protocol (Corman et al., 2020).
Sera
The convalescent human anti-SARS-CoV sera (CSS-2 to CSS-5) stemmed from the serum collection of the national consiliary laboratory for coronavirus diagnostics at Charité, Berlin, Germany or the Robert Koch Institute, Berlin, Germany. All sera were previously tested positive using a recombinant S-based immunofluorescence test (Buchholz et al., 2013). CSS-2 was taken from a SARS patient 3.5 years post onset of disease. CSS-3 and CSS-4 originated from a second SARS patient 24 and 36 days post onset of disease. CSS-5 was collected from a third SARS patient 10 days post onset of disease. Rabbit sera were obtained by immunizing rabbits with purified SARS-S1 protein fused to the Fc portion of human immunoglobulin.
Phylogenetic analysis
Phylogenetic analysis (neighbor-joining tree, bootstrap method with 5,000 iterations, Poisson substitution model, uniform rates among sites, complete deletion of gaps/missing data) was performed using the MEGA7.0.26 software. Reference sequences were obtained from the National Center for Biotechnology Information and GISAID (Global Initiative on Sharing All Influenza Data) databases. Reference numbers are indicated in the figures.
Quantification and Statistical Analysis
One-way or two-way analysis of variance (ANOVA) with Dunnett posttest was used to test for statistical significance. Only p values of 0.05 or lower were considered statistically significant (p > 0.05 [ns, not significant], p ≤ 0.05 [∗], p ≤ 0.01 [∗∗], p ≤ 0.001 [∗∗∗]). For all statistical analyses, the GraphPad Prism 7 software package was used (GraphPad Software).
Data and Code Availability
The study did not generate unique datasets or code.
Acknowledgments
We thank Heike Hofmann-Winkler for discussion, Andrea Maisner for Nipah F and G expression plasmids, Anette Teichmann for technical assistance, and Roberto Cattaneo for plasmid pCG1. We acknowledge the support of the non-profit foundation HTCR, which holds human tissue on trust, making it broadly available for research on an ethical and legal basis. We gratefully acknowledge the authors and the originating and submitting laboratories for their sequence and metadata shared through GISAID, on which this research is based. This work was supported by BMBF (RAPID Consortium, 01KI1723D and 01KI1723A to C.D. and S.P.) and German Research Foundation (DFG) (WU 929/1-1 to N.-H.W.).
Author Contributions
Conceptualization, M.H. and S.P.; Formal analysis, M.H., H.K.-W., M.A.M., and S.P.; Investigation, M.H., H.K.-W., S.S., N.K., T.H., N.-H.W., and M.A.M.; Resources, T.H., S.E., T.S.S., G.H., A.N., M.A.M., and C.D.; Writing – Original Draft, M.H. and S.P.; Writing – Review & Editing, all authors; Funding acquisition, S.P., N.-H.W., and C.D.
Declaration of Interests
The authors declare no competing interests.
Published: March 5, 2020
Contributor Information
Markus Hoffmann, Email: mhoffmann@dpz.eu.
Stefan Pöhlmann, Email: spoehlmann@dpz.eu.
References
- Berger Rentsch M., Zimmer G. A vesicular stomatitis virus replicon-based bioassay for the rapid and sensitive determination of multi-species type I interferon. PLoS ONE. 2011;6:e25858. doi: 10.1371/journal.pone.0025858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Glowacka I., Blazejewska P., Soilleux E., Allen P., Danisch S., Steffen I., Choi S.Y., Park Y., Schneider H. TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J. Virol. 2010;84:10016–10025. doi: 10.1128/JVI.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., Lucas J.M., Nelson P.S., Pöhlmann S., Soilleux E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PLoS ONE. 2012;7:e35876. doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossart K.N., Wang L.F., Flora M.N., Chua K.B., Lam S.K., Eaton B.T., Broder C.C. Membrane fusion tropism and heterotypic functional activities of the Nipah virus and Hendra virus envelope glycoproteins. J. Virol. 2002;76:11186–11198. doi: 10.1128/JVI.76.22.11186-11198.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmann C., Hoffmann M., Lübke A., Nehlmeier I., Krämer-Kühl A., Winkler M., Pöhlmann S. The glycoprotein of vesicular stomatitis virus promotes release of virus-like particles from tetherin-positive cells. PLoS ONE. 2017;12:e0189073. doi: 10.1371/journal.pone.0189073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz U., Müller M.A., Nitsche A., Sanewski A., Wevering N., Bauer-Balci T., Bonin F., Drosten C., Schweiger B., Wolff T. Contact investigation of a case of human novel coronavirus infection treated in a German hospital, October-November 2012. Euro Surveill. 2013;18:20406. [PubMed] [Google Scholar]
- Chan J.F., Yuan S., Kok K.H., To K.K., Chu H., Yang J., Xing F., Liu J., Yip C.C., Poon R.W. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Lienau J., Witzenrath M. [Coronaviruses as the cause of respiratory infections] Internist (Berl.) 2019;60:1136–1145. doi: 10.1007/s00108-019-00671-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y., He L., Zhang Q., Huang Z., Che X., Hou J., Wang H., Shen H., Qiu L., Li Z. Organ distribution of severe acute respiratory syndrome (SARS) associated coronavirus (SARS-CoV) in SARS patients: implications for pathogenesis and virus transmission pathways. J. Pathol. 2004;203:622–630. doi: 10.1002/path.1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr A.R., Channappanavar R., Perlman S. Middle East Respiratory Syndrome: Emergence of a Pathogenic Human Coronavirus. Annu. Rev. Med. 2017;68:387–399. doi: 10.1146/annurev-med-051215-031152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X.Y., Li J.L., Yang X.L., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gierer S., Bertram S., Kaup F., Wrensch F., Heurich A., Krämer-Kühl A., Welsch K., Winkler M., Meyer B., Drosten C. The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J. Virol. 2013;87:5502–5511. doi: 10.1128/JVI.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowacka I., Bertram S., Müller M.A., Allen P., Soilleux E., Pfefferle S., Steffen I., Tsegaye T.S., He Y., Gnirss K. Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J. Virol. 2011;85:4122–4134. doi: 10.1128/JVI.02232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J., Gong E., Zhang B., Zheng J., Gao Z., Zhong Y., Zou W., Zhan J., Wang S., Xie Z. Multiple organ infection and the pathogenesis of SARS. J. Exp. Med. 2005;202:415–424. doi: 10.1084/jem.20050828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B.J., He Y.Q., Liu X.L., Zhuang Z.X., Cheung C.L., Luo S.W., Li P.H., Zhang L.J., Guan Y.J. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science. 2003;302:276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Haga S., Yamamoto N., Nakai-Murakami C., Osawa Y., Tokunaga K., Sata T., Yamamoto N., Sasazuki T., Ishizaka Y. Modulation of TNF-alpha-converting enzyme by the spike protein of SARS-CoV and ACE2 induces TNF-alpha production and facilitates viral entry. Proc. Natl. Acad. Sci. USA. 2008;105:7809–7814. doi: 10.1073/pnas.0711241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M.L., Lely A.T., Navis G., van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Li J., Heck S., Lustigman S., Jiang S. Antigenic and immunogenic characterization of recombinant baculovirus-expressed severe acute respiratory syndrome coronavirus spike protein: implication for vaccine design. J. Virol. 2006;80:5757–5767. doi: 10.1128/JVI.00083-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Müller M.A., Drexler J.F., Glende J., Erdt M., Gützkow T., Losemann C., Binger T., Deng H., Schwegmann-Weßels C. Differential sensitivity of bat cells to infection by enveloped RNA viruses: coronaviruses, paramyxoviruses, filoviruses, and influenza viruses. PLoS ONE. 2013;8:e72942. doi: 10.1371/journal.pone.0072942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Geier M., Marzi A., Krumbiegel M., Peipp M., Fey G.H., Gramberg T., Pöhlmann S. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem. Biophys. Res. Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Hattermann K., Marzi A., Gramberg T., Geier M., Krumbiegel M., Kuate S., Uberla K., Niedrig M., Pöhlmann S. S protein of severe acute respiratory syndrome-associated coronavirus mediates entry into hepatoma cell lines and is targeted by neutralizing antibodies in infected patients. J. Virol. 2004;78:6134–6142. doi: 10.1128/JVI.78.12.6134-6142.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Pyrc K., van der Hoek L., Geier M., Berkhout B., Pöhlmann S. Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proc. Natl. Acad. Sci. USA. 2005;102:7988–7993. doi: 10.1073/pnas.0409465102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X. Lancet; China: 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y., Kuba K., Rao S., Huan Y., Guo F., Guan B., Yang P., Sarao R., Wada T., Leong-Poi H. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nature. 2005;436:112–116. doi: 10.1038/nature03712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata-Yoshikawa N., Okamura T., Shimizu Y., Hasegawa H., Takeda M., Nagata N. TMPRSS2 Contributes to Virus Spread and Immunopathology in the Airways of Murine Models after Coronavirus Infection. J. Virol. 2019;93 doi: 10.1128/JVI.01815-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawase M., Shirato K., van der Hoek L., Taguchi F., Matsuyama S. Simultaneous treatment of human bronchial epithelial cells with serine and cysteine protease inhibitors prevents severe acute respiratory syndrome coronavirus entry. J. Virol. 2012;86:6537–6545. doi: 10.1128/JVI.00094-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Heinlein C., Hackman R.C., Nelson P.S. Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol. Cell. Biol. 2006;26:965–975. doi: 10.1128/MCB.26.3.965-975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Hoffmann M., Pöhlmann S. Functional analysis of potential cleavage sites in the MERS-coronavirus spike protein. Sci. Rep. 2018;8:16597. doi: 10.1038/s41598-018-34859-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber H., Elzayat M.T., Wang L., Graham B.S., Müller M.A., Drosten C., Pöhlmann S., Hoffmann M. Mutations in the Spike Protein of Middle East Respiratory Syndrome Coronavirus Transmitted in Korea Increase Resistance to Antibody-Mediated Neutralization. J. Virol. 2019;93 doi: 10.1128/JVI.01381-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K., Imai Y., Rao S., Gao H., Guo F., Guan B., Huan Y., Yang P., Zhang Y., Deng W. A crucial role of angiotensin converting enzyme 2 (ACE2) in SARS coronavirus-induced lung injury. Nat. Med. 2005;11:875–879. doi: 10.1038/nm1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- Li W., Zhang C., Sui J., Kuhn J.H., Moore M.J., Luo S., Wong S.K., Huang I.C., Xu K., Vasilieva N. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Hulswit R.J.G., Widjaja I., Raj V.S., McBride R., Peng W., Widagdo W., Tortorici M.A., van Dieren B., Lang Y. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA. 2017;114:E8508–E8517. doi: 10.1073/pnas.1712592114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.T., Zhang J.S., Su N., Xu J.G., Wang N., Chen J.T., Chen X., Liu Y.X., Gao H., Jia Y.P. Safety and immunogenicity from a phase I trial of inactivated severe acute respiratory syndrome coronavirus vaccine. Antivir. Ther. (Lond.) 2007;12:1107–1113. [PubMed] [Google Scholar]
- Liu W., Fontanet A., Zhang P.H., Zhan L., Xin Z.T., Baril L., Tang F., Lv H., Cao W.C. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. J. Infect. Dis. 2006;193:792–795. doi: 10.1086/500469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S., Nagata N., Shirato K., Kawase M., Takeda M., Taguchi F. Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 2010;84:12658–12664. doi: 10.1128/JVI.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery V.D., Dinnon K.H., III, Yount B.L., Jr., McAnarney E.T., Gralinski L.E., Hale A., Graham R.L., Scobey T., Anthony S.J., Wang L. Trypsin treatment unlocks barrier for zoonotic bat coronaviruses infection. J. Virol. 2020;94 doi: 10.1128/JVI.01774-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster V.J., Koopmans M., van Doremalen N., van Riel D., de Wit E. A Novel Coronavirus Emerging in China - Key Questions for Impact Assessment. N. Engl. J. Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- Park J.E., Li K., Barlan A., Fehr A.R., Perlman S., McCray P.B., Jr., Gallagher T. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc. Natl. Acad. Sci. USA. 2016;113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.J., Walls A.C., Wang Z., Sauer M.M., Li W., Tortorici M.A., Bosch B.J., DiMaio F., Veesler D. Structures of MERS-CoV spike glycoprotein in complex with sialoside attachment receptors. Nat. Struct. Mol. Biol. 2019;26:1151–1157. doi: 10.1038/s41594-019-0334-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj V.S., Mou H., Smits S.L., Dekkers D.H., Müller M.A., Dijkman R., Muth D., Demmers J.A., Zaki A., Fouchier R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh W.J., Hsiao C.H., Paddock C.D., Guarner J., Goldsmith C.S., Tatti K., Packard M., Mueller L., Wu M.Z., Rollin P. Immunohistochemical, in situ hybridization, and ultrastructural localization of SARS-associated coronavirus in lung of a fatal case of severe acute respiratory syndrome in Taiwan. Hum. Pathol. 2005;36:303–309. doi: 10.1016/j.humpath.2004.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kanou K., Kawase M., Matsuyama S. Clinical Isolates of Human Coronavirus 229E Bypass the Endosome for Cell Entry. J. Virol. 2016;91 doi: 10.1128/JVI.01387-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato K., Kawase M., Matsuyama S. Wild-type human coronaviruses prefer cell-surface TMPRSS2 to endosomal cathepsins for cell entry. Virology. 2018;517:9–15. doi: 10.1016/j.virol.2017.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulla A., Heald-Sargent T., Subramanya G., Zhao J., Perlman S., Gallagher T. A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J. Virol. 2011;85:873–882. doi: 10.1128/JVI.02062-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G., Gosalia D.N., Rennekamp A.J., Reeves J.D., Diamond S.L., Bates P. Inhibitors of cathepsin L prevent severe acute respiratory syndrome coronavirus entry. Proc. Natl. Acad. Sci. USA. 2005;102:11876–11881. doi: 10.1073/pnas.0505577102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Horby P.W., Hayden F.G., Gao G.F. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–473. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO . 2004. Summary of probable SARS cases with onset of illness from 1 November 2002 to 31 July 2003.https://www.who.int/csr/sars/country/table2004_04_21/en/ [Google Scholar]
- WHO . 2020. Novel Coronavirus(2019-nCoV) Situation Report 23.https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200212-sitrep-23-ncov.pdf?sfvrsn=41e9fb78_4 [Google Scholar]
- Wu N.H., Yang W., Beineke A., Dijkman R., Matrosovich M., Baumgärtner W., Thiel V., Valentin-Weigand P., Meng F., Herrler G. The differentiated airway epithelium infected by influenza viruses maintains the barrier function despite a dramatic loss of ciliated cells. Sci. Rep. 2016;6:39668. doi: 10.1038/srep39668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Matsuyama S., Li X., Takeda M., Kawaguchi Y., Inoue J.I., Matsuda Z. Identification of Nafamostat as a Potent Inhibitor of Middle East Respiratory Syndrome Coronavirus S Protein-Mediated Membrane Fusion Using the Split-Protein-Based Cell-Cell Fusion Assay. Antimicrob. Agents Chemother. 2016;60:6532–6539. doi: 10.1128/AAC.01043-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Du L., Liu C., Wang L., Ma C., Tang J., Baric R.S., Jiang S., Li F. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc. Natl. Acad. Sci. USA. 2014;111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Liu C., Du L., Jiang S., Shi Z., Baric R.S., Li F. Two Mutations Were Critical for Bat-to-Human Transmission of Middle East Respiratory Syndrome Coronavirus. J. Virol. 2015;89:9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeager C.L., Ashmun R.A., Williams R.K., Cardellichio C.B., Shapiro L.H., Look A.T., Holmes K.V. Human aminopeptidase N is a receptor for human coronavirus 229E. Nature. 1992;357:420–422. doi: 10.1038/357420a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Vedantham P., Lu K., Agudelo J., Carrion R., Jr., Nunneley J.W., Barnard D., Pöhlmann S., McKerrow J.H., Renslo A.R., Simmons G. Protease inhibitors targeting coronavirus and filovirus entry. Antiviral Res. 2015;116:76–84. doi: 10.1016/j.antiviral.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The study did not generate unique datasets or code.