Abstract
Antimalarial drug resistance is a major public health problem in China. From 2012 to 2015, more than 75% of malaria cases in Shandong Province were P. falciparum returned from Africa. However, molecular marker polymorphisms of drug resistance in imported P. falciparum cases have not been evaluated. In this study, we analyzed polymorphisms of the Pfcrt, Pfmdr1, and Pfkelch13 genes in 282 P. falciparum cases returned from Africa to Shandong between 2012 and 2015. Among the isolates, polymorphisms were detected in codons 74–76 of Pfcrt and 86, 184, 1246 of Pfmdr1, among which K76T (36.6%) and Y184F (60.7%) were the most prevalent, respectively. Six Pfcrt haplotypes and 11 Pfmdr1 haplotypes were identified and a comparison was made on the prevalence of haplotypes among East Africa, West Africa, Central Africa and South Africa. One synonymous and 9 nonsynonymous mutations in Pfkelch13 were detected in the isolates (4.6%), among which a candidate artemisinin (ART) resistance mutation P553L was observed. The study establishes fundamental data for detection of chloroquine resistance (CQR) and ART resistance with molecular markers of the imported P. falciparum in China, and it also enriches the genetic data of antimalarial resistance for the malaria endemic countries in Africa.
Introduction
Malaria is one of the most important parasitic diseases presented in many countries that causing a serious global public health problem. There were approximately 214 million malaria cases and 438,000 deaths worldwide in 2015, of which approximately 88% occurred in Africa1. P. falciparum, the most threatening among five species of malaria parasites associated with human infection, has been mainly responsible for the majority of the morbidity and mortality around the world2. During the past decades, drug resistance of P. falciparum has become a critical obstacle to control and eliminate malaria. Long-term anti-malarial monotherapy is capable of causing the emergence of drug resistant parasite strains and may provide a chance to spread the resistance to sensitive parasite population in other epidemic areas under conditions suitable Anopheles mosquitoes for transmission3. Since the early 1940s, chloroquine (CQ) was continuously used for treatment of malaria in many countries, which has been confirmed one of the most important antimalarial drugs with quick metabolism, good curative effects, and low prices4. However, due to excessive drug usage over the years, the CQ resistance (CQR) of P. falciparum isolates was found initial emerging from Thailand-Cambodia border in Southeast Asia in 1957 and Venezuela-Colombia border in Northern South America in 19595,6, and eventually spread to other countries around the world. In 2006, the World Health Organization (WHO) recommended artemisinin-based combination therapies (ACTs) as the first-line treatment for uncomplicated P. falciparum7. ACTs serve as an important therapeutic method to avoid or defer the development of drug resistance for P. falciparum infection. Unfortunately, P. falciparum resistance to artemisinin (ART), the cornerstone of ACTs, was emergence in western Cambodia and subsequently spreading across several neighboring countries in Greater Mekong Subregion (GMS) of Southeast Asia in recent years8–13. Moreover, the emergence of ART resistant indigenous isolates in Africa has been reported more recently, which would be anxious in the future since the suitable replacement drugs are limited14. Thus, it is urgent to monitor the drug resistant trend of P. falciparum, so as to assess the possibility of reintroducing conventional drugs and also attempt to block the emergence of potential large-scale ART resistant transmission.
Molecular marker detection for parasite’s drug resistance is one of several methods for the surveillance of resistant prevalence and antimalarial efficacy15. The single nucleotide polymorphisms (SNPs) at codons 72, 74, 75 and 76 of P. falciparum chloroquine resistance transporter gene (Pfcrt), and 86, 184, 1034, 1042, and 1246 of P. falciparum multidrug resistance 1 gene (Pfmdr1) have been shown to be associated with parasite’s CQR16,17. Moreover, Pfkelch13, a gene locating on chromosome 13 of P. falciparum and encoding K13-propeller protein, was identified playing a vital role in conferring ART resistance through whole genome sequencing of ART resistance isolate and vitro ring-stage survival assays (RSA0–3h)18. To date, 13 nonsynonymous mutations (P441L, F446I, G449A, N458Y, Y493H, R539T, I543T, P553L, R561H, V568G, P574L, C580Y, A675V) on Pfkelch13 gene of P. falciparum have been reported to be associated with clinical ART resistance that occurred in Southeast Asia19. Among them, Y493H, R539T, I543T, and C580Y were validated ART-resistant mutations, whereas the rest were candidate resistance mutations20.
In China, owing to substantial efforts for strategies and intervention over the past decades, the country has achieved a great success in controlling malaria transmission, with morbidity and mortality dramatically reduced to low levels21. In 2010, the Chinese Government initiated the National Malaria Elimination Program (NMEP), in order to eliminate malaria nationwide by the year 202022. Currently, indigenous malaria parasite was almost absented in majority of regions in China other than some local transmission still occurred in Yunnan Province and Tibet Autonomous Region23. However, due to the intensive commercial intercourse, travelling and migrant laborers, a markedly rise of imported cases in recent years has posed a severe threat to eradicate malaria24. There has been no indigenous malaria patient reported in Shandong Province of China since 2012, whereas the imported cases have increased gradually, especially P. falciparum coming back from Africa was predominant25. Since little is known about the molecular basis of drug resistance of imported P. falciparum in Shandong Province, we investigated polymorphisms and haplotypes distribution of Pfcrt and Pfmdr1, and mutations in Pfkelch13 of P. falciparum isolates returned from Africa in Shandong between 2012 and 2015, in order to accumulate and update baseline data for molecular surveillance linked to antimalarial drug resistance in China.
Results
Epidemiologic profile of cases
Totally 282 uncomplicated P. falciparum cases that returned from 23 countries of Africa to Shandong Province from 2012 to 2015 were enrolled in this study. Among these cases, the majority was coming back from Central Africa (35.1%, 99/282), followed by Southern Africa (30.5%, 86/282), Western Africa (26.6%, 75/282), Eastern Africa (7.8%, 22/282), but no patient was returned from Northern Africa (Table 1). There were 279 male patients and 3 female patients (93:1). The distribution of patients by age ranged from 19 to 60, with 92.6% (261/282) occurred from 20 to 50 years old. The patient’s occupations were mainly consisted of farmers (58.2%, 164/282) and laborers (23.1%, 65/282), and also included business (8.9%, 25/282), house workers (6.0%, 17/282), sailors (1.4%, 4/282), and other unknown occupations (2.5%, 7/282). Before travelling to Africa, all patients received health education conducted by local CDCs, including knowledge about malaria transmission and diagnosis, personal protection, mosquito prevention, and standard treatments. The patients accepted ACTs treatment specified according to the guidelines and regimens for the use of antimalarial drugs in China (2009)26. Clinical features showed that all patients recovered well after they took the therapy and there is no malaria recrudescence through continuous follow-up.
Table 1.
Imported cases returned from regions and countries of Africa.
| Regions and countries | No. of cases |
|---|---|
| Eastern Africa | 22 |
| Sudan | 12 |
| Tanzania | 8 |
| Ethiopia | 2 |
| Western Africa | 75 |
| Nigeria | 34 |
| Ghana | 20 |
| Guinea | 11 |
| Liberia | 3 |
| Sierra Leone | 2 |
| Niger | 2 |
| Ivory Coast | 1 |
| Burkina Faso | 1 |
| Mali | 1 |
| Central Africa | 99 |
| Equatorial Guinea | 77 |
| Republic of Congo | 12 |
| Cameroon | 6 |
| Chad | 3 |
| Gabon | 1 |
| Southern Africa | 86 |
| Angola | 66 |
| Mozambique | 13 |
| Zambia | 4 |
| Malawi | 1 |
| Madagascar | 1 |
| South Africa | 1 |
| Total | 282 |
Polymorphisms of Pfcrt and Pfmdr1 genes
Among the 282 isolates, 279 (98.9%) for Pfcrt gene and 272 (96.5%) for Pfmdr1 gene were successfully sequenced after PCR amplification, whereas 3 isolates of Pfcrt (Equatorial Guinea: n = 2, Nigeria: n = 1) and 10 isolates of Pfmdr1 (Equatorial Guinea: n = 2, Ghana: n = 2, Nigeria: n = 1, Sudan: n = 1, Ethiopia: n = 1, Niger: n = 1) were failed to obtain sequences due to poor quality of DNA. The prevalence of Pfcrt and Pfmdr1 polymorphisms were shown in Table 2. The entire codons 72–76 of Pfcrt were well examined and no polymorphism was found in the position 72 and 73. Of the codons 74, 75 and 76, 36.6% (102/279) of isolates carried polymorphisms and K76T accounting for the same percentage was the most prevalent (36.6%, 102/279). For Pfmdr1, the residues 130, 1034, 1042 and 1109 of all isolates were wild-type alleles. Among the mutant alleles at codons 86, 184 and 1246, polymorphisms were found in 65.4% (178/272) isolates and Y184F with 60.7% (165/272) prevalence was more frequent than the others. In addition, 0.7% (2/272) of the isolates carried D1246Y mutation.
Table 2.
Polymorphisms of Pfcrt and Pfmdr1 in isolates returned from Africa.
| Gene | Codons position | No. of isolates | Prevalence of mutation |
|---|---|---|---|
| Pfcrt (N = 279) | M74I | 100 | 35.8% |
| N75E/D | 100 | 35.8% | |
| K76T | 102 | 36.6% | |
| Pfmdr1 (N = 272) | N86Y | 82 | 30.2% |
| Y184F | 165 | 60.7% | |
| D1246Y | 2 | 0.7% |
Geographic distribution of Pfcrt haplotypes
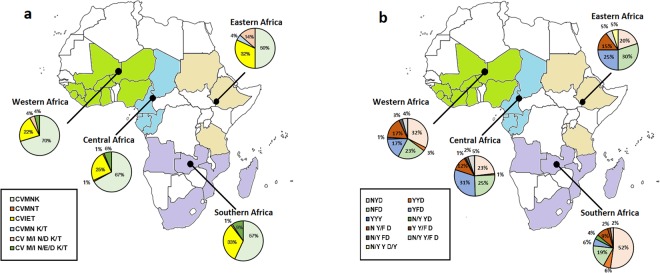
Of the Pfcrt gene, 6 haplotypes coding 72–76 were confirmed, including wild type C72V73M74N75K76 (CVMNK), mutant type CVMNT (mutated amino acids underlined), CVIET, and mixed type CVMN K/T, CV M/I N/D K/T, CV M/I N/E/D K/T, with prevalence of 63.4% (177/279), 0.4% (1/279), 26.9% (75/279), 0.4% (1/279), 2.9% (8/279) and 6.1% (17/279), respectively. Of these, CVMNT and CVMN K/T were only detected once in the isolates. Among the other four haplotypes, CVMNK, CVIET and CV M/I N/D K/T were found in all regions from Africa with no isolate carried CV M/I N/E/D K/T from Eastern Africa. The prevalence of CVMNK in Eastern Africa, Western Africa, Central Africa, and Southern Africa was 50.0% (11/22), 70.3% (52/74), 67.0% (65/97), and 57.0% (49/86), respectively. There was no significant difference among the groups (χ2 = 5.28, P > 0.05). The largest proportion of isolates with CVIET was returned from Southern Africa (32.6%, 28/86), followed by Eastern Africa (31.8%, 7/22), Central Africa (24.7%, 24/97) and Western Africa (21.6%, 16/74), and no significant difference was observed (χ2 = 2.95, P > 0.05). The total proportion of mixed type haplotypes (CVMN K/T, CV M/I N/D K/T, and CV M/I N/E/D K/T) was most occurred in Eastern Africa (18.2%, 4/22), followed by Southern Africa (10.5%, 9/86), Western Africa (8.1%, 6/74), and Central Africa (7.2%, 7/97), and there was no significant difference among the groups (χ2 = 2.81, P > 0.05). The detail information about distribution of Pfcrt haplotypes is shown in Table 3 and Fig. 1a.
Table 3.
Distribution of Pfcrt and Pfmdr1 haplotypes in isolates returned from Africa.
| Gene | Haplotype | Eastern Africa | Western Africa | Central Africa | Southern Africa | Total |
|---|---|---|---|---|---|---|
| Pfcrt | CVMNK | 11 (50.0%) | 52 (70.3%) | 65 (67.0%) | 49 (57.0%) | 177 (63.4%) |
| CVMNT | 0 | 0 | 1 (1.0%) | 0 | 1 (0.4%) | |
| CVIET | 7 (31.8%) | 16 (21.6%) | 24 (24.7%) | 28 (32.6%) | 75 (26.9%) | |
| CVMN K/T | 1 (4.5%) | 0 | 0 | 0 | 1 (0.4%) | |
| CV M/I N/D K/T | 3 (13.6%) | 3 (4.1%) | 1 (1.0%) | 1 (1.2%) | 8 (2.9%) | |
| CV M/I N/E/D K/T | 0 | 3 (4.1%) | 6 (6.2%) | 8 (9.3%) | 17 (6.1%) | |
| Total | 22 | 74 | 97 | 86 | 279 | |
| Pfmdr1 | NYD | 4 (20.0%) | 23 (32.4%) | 22 (23.2%) | 45 (52.3%) | 94 (34.6%) |
| YYD | 0 | 2 (2.8%) | 1 (1.1%) | 5 (5.8%) | 8 (2.9%) | |
| NFD | 6 (30.0%) | 16 (22.5%) | 24 (25.3%) | 16 (18.6%) | 62 (22.8%) | |
| YFD | 5 (25.0%) | 12 (16.9%) | 29 (30.5%) | 5 (5.8%) | 51 (18.8%) | |
| YYY | 0 | 1 (1.4%) | 0 | 0 | 1 (0.4%) | |
| N/Y YD | 0 | 0 | 0 | 3 (3.5%) | 3 (1.1%) | |
| N Y/F D | 3 (15.0%) | 12 (16.9%) | 11 (11.6%) | 8 (9.3%) | 34 (12.5%) | |
| Y Y/F D | 0 | 0 | 1 (1.1%) | 2 (2.3%) | 3 (1.1%) | |
| N/Y FD | 0 | 2 (2.8%) | 2 (2.1%) | 0 | 4 (1.5%) | |
| N/Y Y/F D | 1 (5.0%) | 3 (4.2%) | 5 (5.3%) | 2 (2.3%) | 11 (4.0%) | |
| N/Y Y D/Y | 1 (5.0%) | 0 | 0 | 0 | 1 (0.4%) | |
| Total | 20 | 71 | 95 | 86 | 272 |
Figure 1.
Geographical distribution of Pfcrt (Panel a) haplotypes and Pfmdr1 (Panel b) haplotypes in imported P. falciparum isolates from Africa. Color difference in the map represents the parasites distribution in Africa. No isolate involved in the study are shown in white. The isolates returned from Eastern Africa, Western Africa, Central Africa and Southern Africa are shown in primrose yellow, green, light blue and purple, respectively. Pie charts presenting the frequencies of different haplotypes.
Geographic distribution of Pfmdr1 haplotypes
Totally, 11 haplotypes were identified according to variation of codons 86, 184, and 1246, including wild type N86Y184D1246 (NYD), mutational-types YYD, NFD, YFD, YYY, and mixed type N/Y YD, N Y/F D, Y Y/F D, N/Y FD, N/Y Y/F D, N/Y Y D/Y, accounting for 34.6% (94/272), 2.9% (8/272), 22.8% (62/272), 18.8% (51/272), 0.4% (1/272), 1.1% (3/272), 12.5% (34/272), 1.1% (3/272), 1.5% (4/272), 4.0% (11/272) and 0.4% (1/272), respectively. NFD, YFD and N Y/F D were the top three prevalent of the non-wild haplotypes. The highest percentage of the NYD haplotype was found in Southern Africa, accounting for 52.3% (45/86), followed by Western Africa (32.4%, 23/71), Central Africa (23.2%, 22/95), and Eastern Africa (20.0%, 4/20). There was significant difference among the groups (χ2 = 19.49, P < 0.05). Among the Eastern, Western, Central and Southern areas, the overall mutational type haplotypes accounted for 55.0% (11/20), 43.7% (31/71), 56.8% (54/95) and 30.2% (26/86) respectively, and significant difference among the groups was observed (χ2 = 13.83, P < 0.05). The proportion of total mixed type haplotypes was 25.0% (5/20) in Eastern Africa, 23.9% (17/71) in Western Africa, 20.0% (19/95) in Central Africa, and 17.4% (15/86) in Southern Africa, and there was no significant difference among the groups (χ2 = 1.27, P > 0.05). The detail information about distribution of Pfmdr1 haplotypes is shown in Table 3 and Fig. 1b.
Analysis of Pfkelch13 mutations
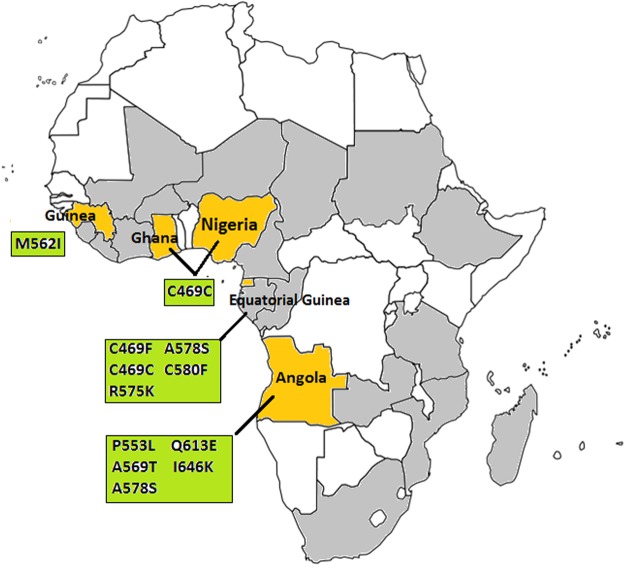
Propeller domain of the P. falciparum Pfkelch13 gene was successful sequenced from all 282 samples and no isolate carried more than one Pfkelch13 mutation. The distribution of Pfkelch13 mutations is shown in Fig. 2. The prevalence of Pfkelch13 mutations was 4.6% (13/282), among which isolates from Equatorial Guinea (1.8%, 5/282) and Angola (1.8%, 5/282) were more frequent than others. Ten different mutant alleles including one synonymous and 9 nonsynonymous were observed, of which C469C, M562I and I646K were unreported before, and C469F and R575K were not previously reported in African isolates (Table 4). Among them, C469C was the most prevalent synonymous mutation (1.1%, 3/282) and A578S was the most prevalent nonsynonymous mutation (0.7%, 2/282), whereas the rest were observed only once. Notably, a candidate resistance mutation P553L was observed in an isolate back from Angola. However, none of the validated ART resistant mutations were observed in the parasites returned from Africa.
Figure 2.
Geographical distribution of Pfkelch13 mutations in imported P. falciparum isolates from Africa. Color difference in the map represents the parasites distribution in Africa. No isolates involved in the study are shown in white. The isolates with no Pfkelch13 mutations are shown in gray. The isolates carried Pfkelch13 mutations are shown in yellow. The Pfkelch13 mutations are shown in green box.
Table 4.
Pfkelch13 mutations in isolates returned from Africa. S, synonymous mutation. NS, nonsynonymous mutations.
| Mutation | Type | Source countries (No. of isolates) |
|---|---|---|
| C469F | NS | Equatorial Guinea (N = 1) |
| C469C* | S | Equatorial Guinea (N = 1), Ghana N = 1), Nigeria (N = 1) |
| P553L | NS | Angola (N = 1) |
| M562I* | NS | Guinea (N = 1) |
| A569T | NS | Angola (N = 1) |
| R575K | NS | Equatorial Guinea (N = 1) |
| A578S | NS | Equatorial Guinea (N = 1), Angola (N = 1) |
| C580F | NS | Equatorial Guinea (N = 1) |
| Q613E | NS | Angola (N = 1) |
| I646K* | NS | Angola (N = 1) |
*Mutation was unreported before.
Discussion
In response to uncomplicated P. falciparum infections, ACTs including oral compound tablets of dihydroartemisinin plus piperaquine (DHA + PQ), artesunate plus amodiaquine (AS + AQ), and ART + PQ were routine regimen for the current treatment in China27. The widespread resistance to CQ, AQ, and the emergence of ART resistant isolates in Africa could attract China’s attention since the resistant parasites would enter the country with increasing migrants. Therefore, understanding the molecular mutation profiles and geographical distribution of drug resistance of P. falciparum is urgent and important for the effective treatment of malaria in China.
Shandong Province is an important coastal economic region located in Eastern China. Historically, Shandong was one of the most severe malaria transmission areas in China, with millions of annual malaria case numbers outbreak in the province during the 1960s and 1970s28. Relying on substantial efforts for anti-malarial campaigns over the decades, the malaria cases in the province decreased sharply and epidemics was well controlled. Nevertheless, there was an increasing trend of the malaria cases presented in Shandong from 2012 to 2015 due to the proportion of imported malaria (100%, 586/586) and the majority of them were P. falciparum cases returning from African countries (79.9%, 468/586)29,30. Since 2009, DHA + PQ and AS + AQ have been applied to against uncomplicated P. falciparum in Shandong as recommended by WHO in China7,26. Nevertheless, little is known about current drug resistance of imported malaria cases. Thus, we assessed polymorphisms of the Pfcrt, Pfmdr1, and Pfkelch13 genes in order to provide genetic data for antimalarial drug resistance.
Polymorphisms in the amino acid positions 72–76 of Pfcrt gene are reliable markers for CQR of P. falciparum parasites, of which K76T mutation is the predominant31. In our study, Pfcrt mutant allele was found together with K76T mutation, which was consistent with above conclusion. For CQR parasites, CVIET and SVMNT are the two main mutant haplotypes prevalent worldwide32,33. The CVIET haplotype has been shown to be predominant in many African countries and is almost the unique haplotype with high frequency in some areas of Africa34. In this study, CVIET (26.9%) was more frequently than other mutant haplotypes and had no significant difference among four regions of Africa, was consistent with above conclusions. Interestingly, CVMNT haplotype was detected in an isolate returned from Equatorial Guinea in this study, which was also observed in 7% of Nigerian isolates and 70.6% of Ghananian isolates previously35,36, suggesting a distinct difference was present in epidemic distribution of CVMNT in Africa. Except mutant types, mixed genotypes of Pfcrt had been detected in 6.6% isolates from Equatorial Guinea37, and then 4.8% mixed types were found in parasites returned from ten countries in Africa38. In present study, 9.3% (26/279) isolates with Pfcrt mixed genotypes were found coming back from 11 source countries in Africa. It further enriched the geographical range of Pfcrt mixed types and also suggested Pfcrt mixed alleles were widely prevalent in African countries. It was known that the isolates with SVMNT haplotype were found most prevalent in South America and Southeast Asia but considered rare in Africa39. This was consistent with our results showing that no isolate carried SVMNT from Africa. However, several recent studies indicated that SVMNT had been detected in parasite strains from Tanzania and Angola. This could be associated with relatively low efficacy of AQ monotherapy in the countries40,41. Therefore, continuous surveillance of SVMNT haplotype is still required for African imported malaria in China. The cessation of CQ for a period of time may lead to the restoration of CQ sensitive parasites. In Malawi, the prevalence of CQR Pfcrt genotype decreased from 85% to 13% during 10 years after withdrawal of CQ42. The same situation also happened to patients from Southern Ethiopia and travelers returned from West and Central African countries43,44. In our samples, the parasites returned from Western Africa and Central Africa carried 70.3% and 67.0% of wild CVMNK haplotype in contrast to 21.6% and 24.7% of mutant CVIET haplotype respectively, which was consistent with above conclusion.
SNPs of Pfmdr1 gene was selected for CQR, and they also had been reported to be associated with regulating drug susceptibility or tolerance to several antimalarials, for example, quinine (QN), mefloquine (MQ), lumefantrine (LU), and even ART45. In this study, Pfmdr1 allelic variants was only observed in codons 86, 184 and 1246. Previous studies suggested that Pfmdr1 N86Y mutation was a potential marker for CQR while Y184F may also play a role in mediating resistance to several antimalarial drugs3,46. Among the isolates, a high frequency of N86Y (30.2%) and Y184F (60.7%) were observed in our study, of which Y184F was more prevalent. It was similar to the previous results in Senegal (14.9% and 71.8%) and Equatorial Guinea (50.3% and 87.3%)37,47. In addition, linkage disequilibrium between K76T and N86Y has been observed in Africa previously48. In this study, both Pfcrt K76T and Pfmdr1 N86Y were detected in 8.9% (25/282) parasite isolates. The correlation of two mutations associated with drug resistance will be of concern in further survey. Considering Pfmdr1 codons 1034, 1042, and 1246, the mutational haplotypes has occurred frequently among CQR parasites in South America, whereas wild haplotype is common in CQR isolates from Africa and Asia35. Interestingly, D1246Y mutation were found in our samples, one reason for this might be ascribed to population flows between Africa and America. For Pfmdr1 wild haplotypes and mutant haplotypes, frequency diversity was observed among four regions of Africa (P < 0.05) respectively. It might be related to diversity of drug pressure and transmission intensity among the countries in Africa.
ACTs are the first-line treatment for P. falciparum in the majority of endemic countries and has been identified as the most successful antimalarial drug over the past 10 years49. Pfkelch13 gene is essential for molecular surveillance of malaria parasites with ART resistance. So far, more than 150 nonsynonymous mutations contracted in Pfkelch13 gene have been reported19. In this study, 9 nonsynonymous Pfkelch13 mutations were observed in the isolates and most of which were returned from Equatorial Guinea and Angola. One possible reason was that samples from two countries were significantly more than others. Previous study reported that R539T mutation was detected in isolates returned from Angola and Equatorial Guinea respectively50. In addition, R539T and C580Y was found in migrant workers returning from Ghana to China36. In our study, no validated ART-resistant Pfkelch13 mutations was observed, the difference could be probably explained by sample sizes. Especially, a candidate ART-resistant mutation P553L was found in an isolate returned from Angola in this study, which also had been detected in Cambodia, Vietnam and West Africa19. These available data suggested that the extensive distribution of low frequent Pfkelch13 nonsynonymous mutations in African P. falciparum population and the emergence of Pfkelch13 mutations in Africa might lead the risk of global resistance transmission. The A578S mutation, which was commonly observed in Africa and several Southeast Asian countries, has been proven to be not related to clinical ART resistance19. In our study, A578S was the most prevalent nonsynonymous mutations among the samples, which was consistent with above results. Notably, one recent report showed a Chinese patient in Jiangsu Province carried ART resistant P. falciparum from Equatorial Guinea, indicating the emergence of indigenous ART resistant isolate in Africa14. Although there is no evidence that ART resistant P. falciparum parasite has emerged in Shandong, the attention should be paid to the increased imported malaria in the province. Therefore, routine molecular surveillance, clinical investigation and field research should be continuously strengthened for awareness of potential emergence of resistance to ACTs from Africa.
In conclusion, our study evaluated polymorphisms and geographic distribution of haplotypes of Pfcrt gene and Pfmdr1 gene in uncomplicated P. falciparum cases imported from Africa to Shandong Province of China. The prevalence of Pfcrt K76T and Pfmdr1 N86Y were still modestly present, indicating the presence of CQR in imported P. falciparum cases. We also detected one synonymous and 9 nonsynonymous mutations in propeller domain of Pfkelch13 gene, among which a candidate ART resistance mutation P553L was observed and 3 mutations were unreported before. Nevertheless, no validated ART resistance mutation of Pfkelch13 gene was found in this study, suggesting no immediate risk to the effect of ART. The study establishes fundamental data for the detection of CQR and ART resistance with molecular markers of the imported P. falciparum in China, and it also enriches the genetic data of drug resistance for the malaria endemic countries in Africa.
Methods
Sample and demographic data collection
Blood samples were obtained from malaria cases who returned from Africa to Shandong Province between 2012 and 2015 prior to antimalarial drug treatment. Demographic data of all cases were recorded, including gender, age, occupation and source countries. The confirmed diagnosis of P. falciparum was performed by microscopic examination of Giemsa-stained thick smears and nested PCR amplifying small-subunit rRNA gene of Plasmodium spp., as described previously51,52. For each specimen, approximately 200 µl finger-prick blood was spotted onto a piece of 3 MM Whatman filter paper. After air dried, blood papers were marked with names, serial numbers and dates, and then stored at −20 °C in individual pouch until DNA extraction.
Ethical approval
This study was reviewed and approved by the Ethics Committee of Shandong Institute of Parasitic Diseases, Shandong Academy of Medical Sciences (Jining, China). All methods were performed in accordance with the relevant guidelines and regulations. The informed consent was obtained from all individual patients or their legal guardians prior to the research.
DNA preparation
Parasitic DNA was isolated from filter paper blots through a QIAamp DNA mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The DNA template was kept at −20 °C until use.
Nested PCR amplification
The known polymorphisms relating to drug resistance at codons 72, 74, 75, 76 of Pfcrt gene and codons 86, 130, 184, 1034, 1042, 1109, 1246 of Pfmdr1 gene, and also mutations on propeller domain of Pfkelch13 gene, were evaluated by nested PCR as described in previous studies18,53,54. The target fragments covering polymorphic sites were as follows: amino acid position 51–83 (a 145 bp portion) for Pfcrt, amino acid position 69–228 (a 526 bp portion) and 1030–1282 (a 799 bp portion) for Pfmdr1, and amino acid position 433–702 (a 849 bp portion) for Pfkelch13. The details of nested PCR primers and conditions are shown in the Supplementary Table S1. The products were analyzed by 1.5% agarose gel electrophoresis stained with SYBR Gold and visualized using a ChemiDoc XRS system (Bio−Rad, Hercules, CA, USA).
DNA sequencing
The successful amplified PCR products were sequenced by the BGI Corporation (Beijing, China). Direct sequencing was carried out through a bigdye terminator v3.1 cycle sequencing kit and ABI prism 3730xl DNA analyzer (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s protocol. The sequences were evaluated by Blast search program on NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to ensure accuracy of PCR amplicons. SNPs of sample sequences were analyzed in Bioedit 7.0 by comparing with reference 3D7 strain PF3D7_0709000 (Pfcrt), PF3D7_0523000 (Pfmdr1) and PF3D7_1343700 (Pfkelch13) from PlasmoDB (http://plasmodb.org/plasmo/). The mixed alleles were determined according to the emergence of two chromatogram peaks at one nucleotide site through the Mutation Surveyor v4.0.0 (SoftGenetics LLC., State College, PA, USA).
Statistical analyses
Data was established using Microsoft Excel 2007 and analyzed by SPSS 19.0 (SPSS Inc., Chicago, IL, USA). The Chi squared test was used to evaluate differences among the groups. A P−value < 0.05 was considered to be statistical significance. The map was created by MapInfo 15.0 (Pitney Bowes, Troy, NY).
Electronic supplementary material
Acknowledgements
This study was supported by Natural Science Foundation of Shandong Province (ZR2014YL036, ZR2016YL019, ZR2017YL003, ZR2018LH016), Projects of Medical and Health Technology Development Program in Shandong Province (2016WS0394), Project of Shandong Academy of Medical Sciences (2017-42), the Innovation Project of Shandong Academy of Medical Sciences, National Natural Science Foundation of China (81702026), China Malaria Project of the Global Fund to Fight AIDS, Tuberculosis and Malaria (201201055). The authors would like to thank the staff members from local CDCs and hospitals in Shandong Province for their assistance in the collection of the samples.
Author Contributions
C.X. and Q.W. made major contributions to writing the manuscript; B.H. conceived and initiated the study; K.Y., H.S., J.L., T.X. and G.Z. participated in sample collection and molecular markers detection; X.K., Y.W., S.Z. and G.Y. participated in the data collection and analyzed the data; K.Y., H.S. and J.K. provided some suggestions for the revision; All authors read and approved the final manuscript.
Data availability
All data generated or analyzed during this study are included in this published article.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Chao Xu and Qingkuan Wei contributed equally.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-31207-w.
References
- 1.World Malaria Report 2015. World Health Organization, Geneva. http://www.who.int/malaria/publications/world-malaria-report-2015/report/en/ (2015).
- 2.Miller LH, Baruch DI, Marsh K, Doumbo OK. Nature. 2002. The pathogenic basis of malaria; pp. 673–679. [DOI] [PubMed] [Google Scholar]
- 3.Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int. 2009;58:201–209. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Telgt DS, van der Ven AJ, Schimmer B, Droogleever-Fortuyn HA, Sauerwein RW. Serious psychiatric symptoms after chloroquine treatment following experimental malaria infection. Ann Pharmacother. 2005;39:551–554. doi: 10.1345/aph.1E409. [DOI] [PubMed] [Google Scholar]
- 5.Harinasuta T, Suntharasamai P, Viravan C. Chloroquine-resistant falciparum malaria in Thailand. Lancet. 1965;286:657–660. doi: 10.1016/S0140-6736(65)90395-8. [DOI] [PubMed] [Google Scholar]
- 6.Moore DV, Lanier JE. Observations on two Plasmodium falciparum infections with an abnormal response to chloroquine. Am J Trop Med Hyg. 1961;10:5–9. doi: 10.4269/ajtmh.1961.10.5. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization. Guidelines for the treatment of malaria. WHO Press: Geneva (2006).
- 8.Noedl H, et al. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med. 2008;359:2619–2620. doi: 10.1056/NEJMc0805011. [DOI] [PubMed] [Google Scholar]
- 9.Dondorp AM, et al. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med. 2009;361:455–467. doi: 10.1056/NEJMoa0808859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hien TT, et al. In vivo susceptibility of Plasmodium falciparum to artesunate in Binh Phuoc Province, Vietnam. Malar J. 2012;11:355. doi: 10.1186/1475-2875-11-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phyo AP, et al. Emergence of artemisinin-resistant malaria on the western border of Thailand: a longitudinal study. Lancet. 2012;379:1960–1966. doi: 10.1016/S0140-6736(12)60484-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kyaw MP, et al. Reduced susceptibility of Plasmodium falciparum to artesunate in southern Myanmar. PLoS One. 2013;8:e57689. doi: 10.1371/journal.pone.0057689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang F, et al. A single mutation in K13 predominates in southern China and is associated with delayed clearance of Plasmodium falciparum following artemisinin treatment. J Infect Dis. 2015;212:1629–1635. doi: 10.1093/infdis/jiv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu F, et al. Emergence of Indigenous Artemisinin-Resistant Plasmodium falciparum in Africa. N Engl J Med. 2017;376:991–993. doi: 10.1056/NEJMc1612765. [DOI] [PubMed] [Google Scholar]
- 15.Barrette, A. & Ringwald, P. Global Report on Antimalarial Drug Efficacy and Drug Resistance 2000–2010. WHO Press: Geneva (2010).
- 16.Golassa L, Enweji N, Erko B, Aseffa A, Swedberg G. High prevalence of pfcrt-CVIET haplotype in isolates from asymptomatic and symptomatic patients in south-central Oromia, Ethiopia. Malar J. 2014;13:120. doi: 10.1186/1475-2875-13-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffing S, et al. pfmdr1 amplification and fixation of pfcrt chloroquine resistance alleles in Plasmodium falciparum in Venezuela. Antimicrob Agents Chemother. 2010;54:1572–1579. doi: 10.1128/AAC.01243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ariey F, et al. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. doi: 10.1038/nature12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ménard D, et al. A worldwide map of Plasmodium falciparum K13-propeller polymorphisms. N Engl J Med. 2016;374:2453–2464. doi: 10.1056/NEJMoa1513137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.World Health Organization. Status report on artemisinin and ACT resistance. http://www.who.int/malaria/areas/drug_resistance/updates/en/ (2016).
- 21.Hu T, et al. Shrinking the malaria map in China: measuring the progress of the National Malaria Elimination Programme. Infect Dis Poverty. 2016;5:52. doi: 10.1186/s40249-016-0146-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ministry of Health, Beijing, China. Action plan of China malaria elimination (2010–2020). http://www.moh.gov.cn/mohbgt/s10788/201005/47529.shtml (2010).
- 23.Feng J, Xiao HH, Xia ZG, Zhang L, Xiao N. Analysis of malaria epidemiological characteristics in the People’s Republic of China, 2004–2013. Am J Trop Med Hyg. 2015;93:293–299. doi: 10.4269/ajtmh.14-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang, L. H. D. Treatment and Management of Imported Malaria Cases. Shanghai Scientific and Technical Publishers: Shanghai, China (2010).
- 25.Xia ZG, Feng J, Zhou SS. Malaria situation in the People’s Republic of China in 2012. Zhongguo Ji Sheng Chong Xue Yu Ji Sheng Chong Bing Za Zhi. 2013;31:413–418. [PubMed] [Google Scholar]
- 26.Ministry of Health, Beijing, China. Antimalarial drug policy in China. http://www.nhfpc.gov.cn/zwgkzt/wsbysj/200907/41610.shtml (2009).
- 27.Ministry of Health, Beijing, China. Antimalarial drug policy in China. http://www.moh.gov.cn/zwgkzt/s9499/201605/68001801f8af435bba987b84891aefc6.shtml (2016).
- 28.Bu XQ, et al. Effects of integrated malaria control measures in Shandong Province, 2010. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2012;24:116–118. [PubMed] [Google Scholar]
- 29.Kong XL, et al. Malaria control and prevention towards elimination: data from an eleven-year surveillance in Shandong Province, China. Malar J. 2017;16:55. doi: 10.1186/s12936-017-1708-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C, et al. Characteristics of Imported Malaria and Species of Plasmodium Involved in Shandong Province, China (2012-2014) Korean J Parasitol. 2016;54:407–414. doi: 10.3347/kjp.2016.54.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fidock DA, et al. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/S1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lakshmanan V, et al. A critical role for PfCRT K76T in Plasmodium falciparum verapamil-reversible chloroquine resistance. EMBO J. 2005;24:2294–2305. doi: 10.1038/sj.emboj.7600681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mehlotra RK, et al. Discordant patterns of genetic variation at two chloroquine resistance loci in worldwide populations of the malaria parasite Plasmodium falciparum. Antimicrob Agents Chemother. 2008;52:2212–2222. doi: 10.1128/AAC.00089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Awasthi G, Satya Prasad GB, Das A. Pfcrt haplotypes and the evolutionary history of chloroquine-resistant Plasmodium falciparum. Mem Inst Oswaldo Cruz. 2012;107:129–134. doi: 10.1590/S0074-02762012000100018. [DOI] [PubMed] [Google Scholar]
- 35.Gbotosho, G. O. et al. Different patterns of pfcrt and pfmdr1 polymorphisms in P. falciparum isolates from Nigeria and Brazil: the potential role of antimalarial drug selection pressure. Am J Trop Med Hyg. 86, 211-213 (2012). [DOI] [PMC free article] [PubMed]
- 36.Feng J, Li J, Yan H, Feng XY, Xia ZG. Evaluation of Antimalarial Resistance Marker Polymorphism in Returned Migrant Workers in China. Antimicrob Agents Chemother. 2015;59:326–30. doi: 10.1128/AAC.04144-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li J, et al. Molecular mutation profile of Pfcrt and Pfmdr1 in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Infect Genet Evol. 2015;36:552–556. doi: 10.1016/j.meegid.2015.08.039. [DOI] [PubMed] [Google Scholar]
- 38.Zhou RM, et al. Molecular mutation profile of pfcrt in Plasmodium falciparum isolates imported from Africa in Henan province. Malar J. 2016;15:265. doi: 10.1186/s12936-016-1306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Awasthi G, Das A. Genetics of chloroquine-resistant malaria: a haplotypic view. Mem Inst Oswaldo Cruz. 2013;108:947–961. doi: 10.1590/0074-0276130274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alifrangis M, et al. Occurrence of the Southeast Asian/South American SVMNT haplotype of the chloroquine-resistance transporter gene in Plasmodium falciparum in Tanzania. J Infect Dis. 2006;193:1738–1741. doi: 10.1086/504269. [DOI] [PubMed] [Google Scholar]
- 41.Gama BE, et al. Plasmodium falciparum isolates from Angola show the StctVMNT haplotype in the pfcrt gene. Malar J. 2010;9:174. doi: 10.1186/1475-2875-9-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kublin JG, et al. Reemergence of chloroquine-sensitive Plasmodium falciparum malaria after cessation of chloroquine use in Malawi. J Infect Dis. 2003;187:1870–1875. doi: 10.1086/375419. [DOI] [PubMed] [Google Scholar]
- 43.Mekonnen SK, et al. Return of chloroquine-sensitive Plasmodium falciparum parasites and emergence of chloroquine-resistant Plasmodium vivax in Ethiopia. Malar J. 2014;13:244. doi: 10.1186/1475-2875-13-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gharbi M, et al. Longitudinal study assessing the return of chloroquine susceptibility of Plasmodium falciparum in isolates from travellers returning from West and Central Africa, 2000–2011. Malar J. 2013;12:35. doi: 10.1186/1475-2875-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kavishe RA, et al. Surveillance of artemether-lumefantrine associated Plasmodium falciparum multidrug resistance protein-1 gene polymorphisms in Tanzania. Malar J. 2014;13:264. doi: 10.1186/1475-2875-13-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dokomajilar, C., Nsobya, S. L., Greenhouse, B., Rosenthal, P. J. & Dorsey, G. Selection of Plasmodium falciparum pfmdr1 alleles following therapy with artemether-lumefantrine in an area of Uganda where malaria is highly endemic. Antimicrob Agents Chemother. 50, 1893–1895 (2006). [DOI] [PMC free article] [PubMed]
- 47.Wurtz N, et al. Role of Pfmdr1 in in vitro Plasmodium falciparum susceptibility to chloroquine, quinine, monodesethylamodiaquine, mefloquine, lumefantrine, and dihydroartemisinin. Antimicrob Agents Chemother. 2014;58:7032–7040. doi: 10.1128/AAC.03494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adagu IS, Warhurst DC. Plasmodium falciparum: linkage disequilibrium between loci in chromosomes 7 and 5 and chloroquine selective pressure in Northern Nigeria. Parasitology. 2001;123:219–224. doi: 10.1017/S0031182001008344. [DOI] [PubMed] [Google Scholar]
- 49.Chen C. Development of antimalarial drugs and their application in China: a historical review. Infect Dis Poverty. 2014;3:9. doi: 10.1186/2049-9957-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang CY, et al. Polymorphisms of Plasmodium falciparum k13-propeller gene among migrant workers returning to Henan Province, China from Africa. BMC Infect Dis. 2017;17:560. doi: 10.1186/s12879-017-2634-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.World Health Organization. Center for Disease Control. Basic MalariaMicroscopy: Tutor’s guide. WHO Press: Geneva2010.
- 52.Snounou G, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61:315–320. doi: 10.1016/0166-6851(93)90077-B. [DOI] [PubMed] [Google Scholar]
- 53.Djimdé A, et al. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 54.Li J, et al. High prevalence of pfmdr1 N86Y and Y184F mutations in Plasmodium falciparum isolates from Bioko Island, Equatorial Guinea. Pathog Glob Health. 2014;108:339–343. doi: 10.1179/2047773214Y.0000000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article.