Abstract
We consider the problem of comparing the gene expression levels of cells grown under two different conditions using cDNA microarray data. We use a quality index, computed from duplicate spots on the same slide, to filter out outlying spots, poor quality genes and problematical slides. We also perform calibration experiments to show that normalization between fluorescent labels is needed and that the normalization is slide dependent and non-linear. A rank invariant method is suggested to select non-differentially expressed genes and to construct normalization curves in comparative experiments. After normalization the residuals from the calibration data are used to provide prior information on variance components in the analysis of comparative experiments. Based on a hierarchical model that incorporates several levels of variations, a method for assessing the significance of gene effects in comparative experiments is presented. The analysis is demonstrated via two groups of experiments with 125 and 4129 genes, respectively, in Escherichia coli grown in glucose and acetate.
INTRODUCTION
Although cDNA microarrays have been used for global monitoring of gene expression in many areas of biomedical research (1), methods for analysis of the resulting data are only beginning to be addressed systematically (2–7). We have performed a series of calibration and comparative experiments to address several important issues in data analysis and study design of microarray experiments. In each calibration experiment we purified total RNA from Escherichia coli cells and divided the sample into two aliquots for labeling by Cy3 and Cy5. The two separately labeled samples were then pooled and subdivided into hybridization solutions for hybridization to multiple slides. In the first group of experiments each slide had 125 E.coli genes multiply spotted (4 spots/gene) on it, while in the second each slide had 4129 genes singly spotted. The first and second groups of experiments will be called the 125 and 4129 gene projects, respectively, hereafter. Several levels of replication are embedded in the design of these calibration experiments and the resulting data provide information on the relative importance of variations due to spots, labels and slides. Based on this information, we formulate an approach to the analysis of comparative experiments where the samples to be compared are differentially labeled. The main components are as follows. (i) Detect and filter out poor quality genes on a slide using measurements from multiple spots. This procedure is not applicable in singly spotted designs. (ii) Perform slide-dependent non-linear normalization of the log ratios of the two channels. (iii) Apply hierarchical model-based analysis to the normalized log ratio scale, where assessment of the significance of gene effects are aided by statistical information obtained from calibration experiments, if they are available. Details of the experiments are given below and the analysis methodology is developed, justified and illustrated. A discussion of other important issues, such as why a two label design is useful and whether gene–label interaction is an important consideration, is also provided.
MATERIALS AND METHODS
Preparation of the DNA array
In the 125 gene project, to ensure uniform quality and quantity of the DNA probes, we constructed a gene library consisting of 125 genes each cloned into pBluescript II KS+ (Stratagene, La Jolla, CA) as previously reported (8,9). These genes are involved in various aspects of E.coli physiology, including glycolysis, the TCA cycle, the pentose phosphate pathway, fermentation pathways, the heat shock response, major biosynthetic pathways and the respiratory system. The gene probes used in microarray construction were obtained by PCR amplifying the inserted genes using pBluescript II KS+-specific primers (Genosys, The Woodlands, TX), 5′-GGCCGCTCTAGAACTAGTGGAT-3′ and 5′-CTCGAGGTCGACGGTATCGATA-3′. PCR products were precipitated with ethanol and redissolved in 15 µl of 350 mM sodium bicarbonate/carbonate buffer, pH 9.0. Each gene was spotted four times on a slide to analyze the reliability and variability. In the 4129 gene project we performed the PCR reactions using Genosys E.coli ORFmers (the entire genome of E.coli) and an Eppendorf MasterTaq kit (Westbury, NY). Among 4290 primers, 161 failed to make products or proper sized products. The 4129 PCR products, representing 96% of the predicted open reading frames (10), were precipitated with propanol twice and then dissolved in 10 µl of 350 mM sodium bicarbonate/carbonate buffer, pH 9.0. They were arrayed with single spotting on each slide. All resulting slides with DNA probes underwent post-processing according to the protocol suggested by Eisen and Brown (11).
RNA purification and labeling
Escherichia coli strain MC4100 [F– araD139 (argF-lac) U169 rpsL150 relA1 flb5301 deoC1 ptsF25 rbsR] was cultured in shake flasks using M9 minimal medium (12) containing either 0.5% glucose or acetate as carbon source supplemented with 125 mg/l (w/v) arginine. When the optical density of the cell reached 0.4–0.6 at 550 nm total RNA was purified from 1 × 109 cells using the RNeasy Midi kit from Qiagen (Valencia, CA). The resulting RNA solution was incubated at 37°C with 100 U DNase (Gibco BRL, Rockville, MD) and 40 U RNasin RNase inhibitor (Promega, Madison, WI) for 30 min, extracted with phenol/chloroform and then precipitated with ethanol. After dissolution in 10–20 µl of RNase-free water, 30 µg total RNA was labeled with either Cy3 or Cy5 during reverse transcription. The reverse transcription cocktail included 200 U Superscript RNase H– reverse transcriptase (Gibco BRL), E.coli gene-specific C-terminal primers (Genosys), 0.5 mM dATP, dTTP and dGTP, 0.2 mM dCTP and 0.1 mM Cy3- or Cy5-labeled dCTP (Amersharm Pharmacia, Piscataway, NJ). After reverse transcription the RNA was degraded by adding 5 µl of 1 N NaOH and incubating at 65°C for 40 min. The resulting cDNA, labeled with either Cy3 or Cy5, was diluted with 60 µl of TE buffer, pH 8.0, and then mixed together. The labeled cDNA mixture was then concentrated to 1–2 µl using Micron-50 (Millipore, Bedford, MA).
Hybridization and scanning
The concentrated Cy3- and Cy5-labeled cDNA was resuspended in 10 µl of hybridization solution, consist of 50% formamide, 3× SSC, 1% SDS, 5× Denhardt’s solution, 0.1 mg/ml salmon sperm DNA and 0.05 mg/ml yeast total RNA. Hybridization solution without 5× Denhardt’s solution was also used for comparison. The labeled cDNA was denaturated at 95°C for 3 min then quickly chilled on ice. The cDNA was then placed on the slide and covered by a coverslip. The slide was assembled with a hybridization chamber (Corning, Charlotte, NC) and hybridized for 14–20 h at 42°C. The hybridized slide was washed in 2× SSC, 0.1% SDS for 5 min at room temperature and then 0.2× SSC for 5 min prior to scanning.
After drying the hybridized slides were scanned with an Affymetrix 418 scanner (Santa Clara, CA) and the scanned images analyzed with the software program Imagene (Biodiscovery, Santa Monica, CA). The median intensities of spot areas were calculated and imported into the program S-Plus (MathSoft, Cambridge, MA).
Description of experiments
We performed four calibration experiments and two comparative experiments in the 125 gene project, two calibration and two comparative ones in the 4129 gene project. Calibration experiments used the same mRNA pool divided into two aliquots and labeled separately with two different dyes in order to investigate variations in this technology. Some calibration experiments used genes from E.coli grown in acetate, while the others used E.coli grown in glucose. The comparative experiments labeled mRNA from E.coli grown in acetate with Cy3 and mRNA from E.coli grown in glucose with Cy5. Different slides in the same experiment were hybridized with the same pool of labeled cDNA and different experiments in the same project redid the whole experiment with the same pool of mRNA.
We will use C, R and S to denote the calibration experiment, comparative (real) experiment and slide, respectively, and suffix numbers to indicate the sequence in the two projects. For example, C3S2 indicates slide 2 in the third calibration experiment and R1S2 slide 2 in the first comparative experiment. Some slides did not use Denhardt’s solution during hybridization while others did. Detailed information concerning experimental design is listed in Table 1.
Table 1. Experimental design of the two projects.
| |
|
Slides in the experiment |
Samples in Cy3 |
Samples in Cy5 |
Denhardt’s solution |
| 125 gene project | Calibration | C1S1–C1S2 | Acetate | Acetate | All slides |
| C2S1–C2S4 | Glucose | Glucose | None | ||
| C3S1–C3S2 | Glucose | Glucose | C3S1 | ||
| C4S1–C4S3 | Glucose | Glucose | C4S1–C4S2 | ||
| Comparative | R1S1–R1S2 | Acetate | Glucose | All slides | |
| R2S1∼R2S2 | Acetate | Glucose | All slides | ||
| 4129 gene project | Calibration | C1S1–C1S2 | Acetate | Acetate | All slides |
| C2S1–C2S2 | Glucose | Glucose | All slides | ||
| Comparative | R1S1–R1S2 | Acetate | Glucose | All slides | |
| R2S1–R2S2 | Acetate | Glucose | All slides |
RESULTS AND DISCUSSION
Outline of analysis procedure
The steps of the proposed analysis are herein briefly described. The
motivation and justification of each step will be given in subsequent
sub-sections. To analyze a calibration experiment: (i) compute a
quality measure for each gene and perform quality filtering; (ii)
denote Mpgse = log(Cy5pgse/Cy3pgse)
and Apgse = [log(Cy5pgse) + log(Cy3pgse)]/2
(base 10) of each spot on the slide where gene g = 1,
2, … , G, spot p = 1,
2, … , P, slide s = 1,
2, … , S and experiment e = 1,
2, … , E. Fit a (slide-specific) normalization
curve ![]() =
= ![]() (A) (3); (iii)
compute the normalized log ratios (base 10)
(A) (3); (iii)
compute the normalized log ratios (base 10) ![]() pgse = Mpgse –
pgse = Mpgse – ![]() pgse.
Average the log ratios from the multiple spots of the genes to obtain
the estimated gene effect
pgse.
Average the log ratios from the multiple spots of the genes to obtain
the estimated gene effect ![]() gse = meanp(
gse = meanp(![]() pgse).
pgse).
To analyze comparative experiments we also first performed quality filtering. Then we used a ‘rank invariant’ method (below and Supplementary Material) to select a subset of genes to be used as the basis for constructing the normalization curves in step 2. After normalization we assessed the significance of expression of each gene from the normalized log ratios in comparative experiments using a hierarchical linear model. The log ratios obtained in the analysis of calibration experiments were used to construct prior distributions of the between-slide and between-experiment variance components in this model (below and Supplementary Material).
Quality filtering using multiple spots
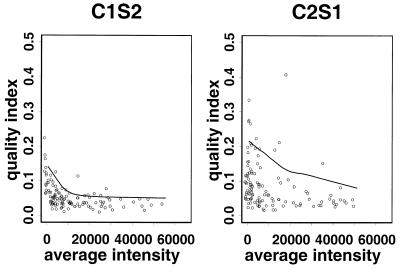
Multiple spotting of target DNA on a slide provides a means to assess the quality of data for a gene on that slide (6). Suppose that each gene is spotted p times on the slide (p = 4 in our 125 gene project). For each spot the ratio of Cy3 and Cy5 intensities was first calculated as m = Cy5/Cy3. We denote by CV the coefficient of variation (i.e. standard deviation divided by the mean) of the set of ratios m1, m2, … , mp on the multiple spots. The quality of data on the expression level of each gene is inversely related to its CV. Figure 1 shows the CV versus mean intensity (average of Cy3 and Cy5 signals) on slides in the 125 gene project.
Figure 1.
Quality index (CV) versus average intensity (Cy5i + Cy3i)/2 in the 125 gene project. The curve indicates the 10th upper percentile in the moving window containing the 50 nearest genes. Genes with a quality index (CV) larger than this curve will be filtered out. Only slides C1S2 and C2S1 are shown here. Genes with a low CV have high agreement in duplicate spots, hence representing high experiment quality. Thus slide C1S2 shows higher quality than slide C2S1.
In Figure 1 we mark all genes having CV values larger than a threshold as poor quality data by a windowing procedure. For each gene we construct a windowing subset by selecting 50 genes whose mean intensities are closest to this gene. If the CV of this gene is within the top 10% among genes in its windowing subset then we regard the data on this gene as unreliable. The curves in Figure 1 show the thresholds used to filter unreliable data. Data from both calibration and comparative experiments in the 125 gene project were filtered using this approach. Following the convention of Dudoit et al. (3), we drew a so-called M–A plot for the initial investigation where M = log(Cy5/Cy3) represents the log ratio of the two dyes and A = [log(Cy5) + log(Cy3)]/2 is the average logarithmic intensity. The plot is actually a 45° rotation and rescaling of the log intensity plot of Cy5 and Cy3. M–A plots of the remaining data for two slides after quality filtering are shown in Figure 2.
Figure 2.
M–A plot for the 125 gene project where M represents the log ratio of two dyes and A the averaged logarithmic intensity. Only slides C1S1 and R1S1 are shown here.
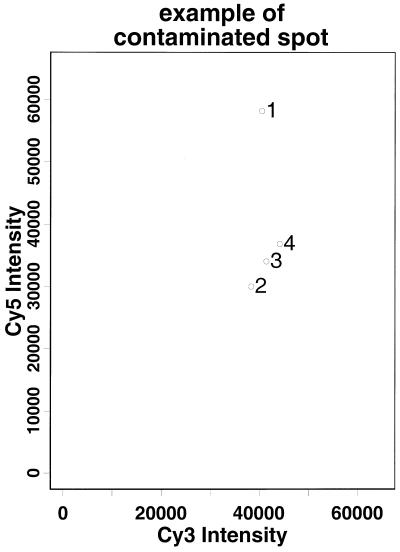
When a gene failed to pass this quality filter we attempted on occasion to salvage information by eliminating the most outlying spot and then recomputed the CV of the intensity ratios of the remaining spots associated with this gene. For example, the intensity ratio of spot 1 in Figure 3 is more than 23 SD from the mean of intensity ratios of the remaining three spots. After removing spot 1 the CV for this gene dropped to 1/10 of its original value. Thus spot 1 is likely to be a contaminated spot, but the remaining spots are still reliable and can be used in subsequent analyses. If, on the other hand, deleting any of the extreme spots does not lead to a significant reduction in CV, then most of the spots of this gene may have been contaminated and we will have to remove this gene from further analysis.
Figure 3.
Cy5 intensity versus Cy3 intensity of the aceE gene on slide C2S2 for the 125 gene project. Spot 1 is a contaminated spot.
Besides screening genes with unreliable data, the CV values can also be used to compare the quality of different slides and different experiments. For example, in analyzing the first two calibration experiments and the comparative experiment in the 125 gene project we found that C2S1–C2S4 are of much poorer quality as compared to R1S1–R1S2, R2S1–R2S2 and C1S1–C1S2 (Fig. 1). Since Denhardt’s solution was used in both the comparative experiments and in the first but not second calibration experiment, we suspect that this might be an explanation. To verify this we performed third and fourth calibrations. It turned out that slides C3S1 and C4S1–C4S2 (with Denhardt’s solution) were of better quality compared to slides C3S2 and C4S3 (without Denhardt’s solution) in the same experiments. This confirms that using Denhardt’s solution can greatly improve experiment quality. Thus multiple spotting can provide useful information on data quality. It allows us to perform quality filtering, i.e. removal of outlying spots and unreliable genes and identification of problematical slides.
Calibration experiment and systematic effects
Calibration experiment. We performed calibration experiments in which the same sample was labeled with both fluorescent dyes. We divided the same pool of RNA, which was extracted from the same E.coli culture, into two aliquots. The two aliquots were separately reverse transcribed in the presence of either Cy3–dCTP or Cy5–dCTP. The two separately labeled cDNA solutions were hybridized to the same slide. The calibration experiment provides a control to investigate possible systematic effects, such as slide effects, dye effects and gene–label interactions, in this technology. The information on non-systematic variation after normalization in calibration experiments can be used to infer the expression level in comparative experiments (see below).
Slide effect. Different slides introduce variations in both hybridization and imaging. Factors that affect hybridization include the amount of probe DNA immobilized on the slide during array fabrication, the amount of labeled cDNA added to the slide and the local environment in each hybridization chamber. During imaging the background noise on the slide and the local curvature of the surface may affect the scanner reading. Confocal scanners are particularly sensitive to focus. The effect of all these factors on intensity measurement is defined as the slide effect. To evaluate the significance of the slide effect we can examine hybridization of a single labeled cDNA pool to different slides. We compared the scattering of logarithmic Cy3 measurements in C1S1 versus C1S2 and the scattering of logarithmic Cy3 versus Cy5 in C1S1. The scattering was more severe across different slides as compared to that across the two dyes. This finding supports the common practice of using a two label design in microarray experiments.
The within-slide variation, which is not a focus of this paper, can also be large. Examples include areas of contamination, high background or uneven cDNA hybridized on the slide surface. In experiments using multiple pins to immobilize probe DNA the pin-to-pin variation can be notable. We used a single pin in the 125 gene project and four pins in the 4129 gene project. The pin-to-pin variation appears to be negligible in our experiments. Analyses concerning notable pin-to-pin variation experiments are comprehensively discussed in Dudoit et al. (3).
Label effect and normalization in calibration. The most commonly used fluorescent dyes, Cy3 and Cy5, are relatively unstable. In addition, these dyes may differentially influence incorporation efficiencies during labeling, have different quantum efficiencies and are detected by the scanner with different efficiencies. The effect of these factors on intensity measurements is defined as the label effect, which is accounted for by the normalization curve in our proposed analysis.
To study the label effect we drew M–A plots in calibration experiments. Because the two cDNA solutions were from the same pool of RNA in calibration, the scanner reading from the Cy3 channel should be identical to that from the Cy5 channel if label effects are negligible. In this ideal case the M–A plot should scatter around the line M = 0. Figure 4 shows the M–A plots after quality filtering for C1S1–C1S2 and C4S1–C4S2 for the 125 gene project. It shows that normalization is needed to account for the label effect. Another notable feature is that the normalization is slide dependent. When the same batches of labeled cDNA were hybridized to a different slide, the M–A data showed a different correlation pattern (Fig. 4, crosses versus open circles). It suggests that there is no universal normalization curve.
Figure 4.
M–A plot of two slides in the same calibration experiment. The upper plot shows different patterns of M–A plot on slides C1S1 (open circles) and C1S2 (crosses) for the 125 gene project. The lower M–A plot for calibration 4 shows the same situation. Thus the normalization curve is slide dependent and should be estimated and applied within the same slide.
Our normalization procedures basically followed Dudoit et al. (3). First we fitted ![]() =
= ![]() (A) to each slide in the calibration experiment.
Fitting can be done by the built-in Lowess function in S-Plus (13). Then the normalized log ratio is
computed by
(A) to each slide in the calibration experiment.
Fitting can be done by the built-in Lowess function in S-Plus (13). Then the normalized log ratio is
computed by ![]() = M –
= M – ![]() .
Note that this normalization procedure is non-linear. The need for
non-linear normalization is also noted in Affymetrix oligonucleodtide
microarray analysis (14). The normalized
log ratio
.
Note that this normalization procedure is non-linear. The need for
non-linear normalization is also noted in Affymetrix oligonucleodtide
microarray analysis (14). The normalized
log ratio ![]() can also be expressed as log(KA × Cy5/Cy3), which shows the
multiplicative nature of the intensity-specific scaling factor.
A variation of this normalization procedure when applied to comparative
experiments is discussed below.
can also be expressed as log(KA × Cy5/Cy3), which shows the
multiplicative nature of the intensity-specific scaling factor.
A variation of this normalization procedure when applied to comparative
experiments is discussed below.
Gene–label interaction. The use of two labels may also introduce gene–label interactions. For example, Cy3–dCTP may be preferentially incorporated into a specific sequence, relative to Cy5–dCTP. If such an interaction exists certain genes will always show higher intensity in one of the channels, even under non-differential expression conditions and after normalization. In such a case the normalized log ratios on different slides in the calibration experiment will be correlated and these correlations can be used to detect gene–label interactions. Table 2 shows that except for C1S1–C1S2 in the 125 gene project the residuals are poorly correlated between different slides in both the 125 and 4129 gene projects. Theoretically some degree of gene–label interaction may exist. However, this interaction appears to be insignificant in magnitude compared to other sources of variation in the present experiment.
Table 2. Correlation of log ratios of paired slides in calibration experiments.
| 125 gene project |
|
|
|
|
4129 gene project |
|
|
|
|
| |
C1S1 |
C1S2 |
C3S1 |
C4S1 |
C4S2 |
C1S1 |
C1S2 |
C2S1 |
C2S2 |
| C1S1 | 1.00 | 0.84 | 0.12 | 0.31 | –0.13 | 1.00 | 0.21 | 0.20 | 0.12 |
| C1S2 | 0.84 | 1.00 | 0.07 | 0.37 | –0.17 | 0.21 | 1.00 | 0.17 | 0.13 |
| C2S1 | 0.20 | 0.17 | 1.00 | 0.31 | |||||
| C2S2 | 0.12 | 0.13 | 0.31 | 1.00 | |||||
| C3S1 | 0.12 | 0.07 | 1.00 | 0.17 | –0.21 | ||||
| C4S1 | 0.31 | 0.37 | 0.17 | 1.00 | 0.36 | ||||
| C4S2 | –0.13 | –0.17 | –0.21 | 0.36 | 1.00 |
A high correlation shows the possibility of gene–label interactions. Except for C1S1 and C1S2 in the 125 gene project, the gene–label interactions are not significant.
Normalization procedure in comparative experiments
In a comparative microarray experiment two differentially expressed mRNA pools are separately labeled with Cy3 or Cy5 and co-hybridized to the same slide. As discussed above, the label normalization function is non-linear and slide dependent. To perform label normalization in a comparative experiment we have to identify a sufficient number of non-differentially expressed genes on each slide and use them to construct a normalization curve.
One solution to this problem is to apply predetermined ‘housekeeping’ genes, which are biologically assumed to be non-differentially expressed genes in the experiments. Note that if the number of predetermined housekeeping genes is small or their intensities do not cover a range of different intensity levels this approach may not provide a good fit for non-linear normalization curves. Also, the expression levels of housekeeping genes can exhibit natural variability. Here we use a rank invariant selection (15) to achieve this goal. The ranks of Cy3 and Cy5 intensities of each gene on the slide are separately computed. For a given gene if the ranks of Cy3 and Cy5 intensities differ by less than a threshold value d and the rank of the averaged intensity is not among the highest l ranks or lowest l ranks this gene is classified as a non-differentially expressed gene. A threshold value of 5 for both d and l was used in the 125 gene project. In the 4129 gene project the larger number of genes allowed us to use a more sophisticated iterative selection scheme (15). In each iteration the threshold for rank difference was determined by the number of selected genes (i.e. genes that had been selected in the last stage) multiplied by a predetermined percentage p. The threshold for rank averaged intensity l is only applied in the first iteration. The iteration stops when the remaining set of genes does not decrease after selection. We use p = 0.02 and l = 25 for the 4129 gene project. Figure S1 in Supplementary Material shows the selection results using the non-iterative and iterative rank invariant methods with different p. It indicates that the iteration procedure helps to select more conserved sets of genes.
This method is based on the assumption that if a gene is up-regulated its intensity rank in one channel, say Cy5, should be significantly higher than the rank in the other, and vice versa. This method may fail in some extreme cases where a majority of the genes are up-regulated (or down-regulated) to the same extent. However, if there are a large number of non-differentially expressed genes, as in the case of most cDNA microarray experiments, this method will work well.
After selecting non-differentially expressed genes and fitting a normalization curve (using the Lowess procedure in S-Plus; 13) in the 4129 gene project we extrapolated the normalization curve to normalize genes with extremely high or low intensities. The extrapolation is based on the 50 genes with the highest and lowest average log intensity ranks in the selected set of non-differentially expressed genes. Detailed illustrations of iterative selection and curve extrapolation are provided in Supplementary Material. Figure 5 shows the extrapolated Lowess curve for M–A plots in comparative experiments for the 4129 gene project.
Figure 5.
Normalization curve for M–A plots in comparative experiments for the 4129 gene project. The darker points are genes of the rank invariant set selected in an iterative manner. (P = 0.02)
Hierarchical model and assessment of gene expression
Since the quality filtering step in the 125 gene experiment identified slides C2S1–C2S4, C3S2 and C4S3 as having poor quality, we used only C1S1–C1S2, C3S1 and C4S1–C4S2 for further analysis.
We used a Bayesian approach to incorporate prior knowledge generated from calibration experiments into the statistical analysis. The prior knowledge is used to construct prior distributions of unobserved parameters. The posterior distribution of the desired parameters is then computed to represent the combined information on the parameters from the observed data and prior distribution.
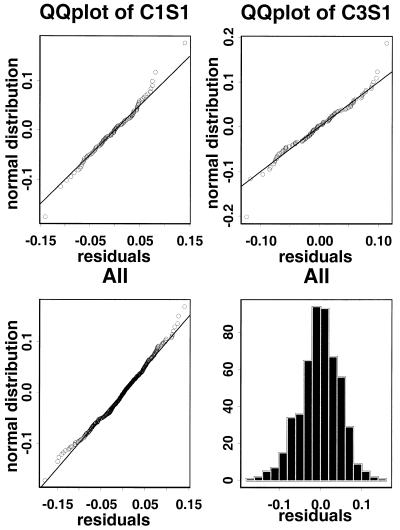
Figure 6 presents the distributions of the normalized log ratios (pooled across all genes) on slides in the calibration experiments for the 125 gene project. The distributions are centered and normal-like and the distributions are very consistent across slides in different experiments. In particular, the distribution is condition independent. Although slides C1S1–C1S2 used one condition (E.coli grown in acetate) and slides C3S1 and C4S1–C4S2 used another (E.coli grown in glucose), the corresponding gene effect distributions were similar. Thus we will incorporate this prior knowledge in the statistical analysis.
Figure 6.
QQ plots and histograms of normalized log ratios in calibration experiments for the 125 gene project. There are ∼100 genes on each slide after quality filtering. The distributions of normalized log ratios are centered, normal-like and consistent across slides. Thus the distributions will provide good prior information for comparative experiments.
We developed the following hierarchical linear model to assess the gene expression level. Denote by ygse the normalized log ratios of gene g, slide s and comparative experiment e. We recognize that ygse is affected by the slide effect and uncontrollable variation between the different bacterial cultures used in different experiments. For each experiment (culture) ygse is a sampling from a normal distribution of the slide effect within the same culture. Thus ygse ∼ N(µge,τg2), where µge is the mean among different slides within this culture and τg2 is the variance in the slide effect distribution for gene g. Furthermore, the within-experiment mean, µge, is in turn a sampling from a normal distribution of culture variation. Thus µge ∼ N(θg,σg2), where θg measures the true log-fold change for gene g and σg2 is the variance between bacterial cultures. Note that only ygse are observed data while τg2, σg2 and θg are unobserved parameters. Under this model θg is the unknown parameter of interest and the derived posterior distribution of θg is used to assess the expression level of gene g. If gene g is non-differentially expressed then θg is distributed around 0. Intuitively, to declare a gene differentially expressed means that ygse deviate from 0 in the same direction and that the deviations are large compared to the magnitude of the posterior distribution of τg2 and σg2.
We give details of the construction of prior distributions of τg2 and σg2 and the statistical test of homogeneity of τg2 in Supplementary Material. A variation of this method when calibration experiments are not available is also discussed. Since the posterior distributions of the parameters do not have a closed form solution, a Markov chain Monte Carlo method (MCMC) (16) was used to simulate the desired posterior distributions.
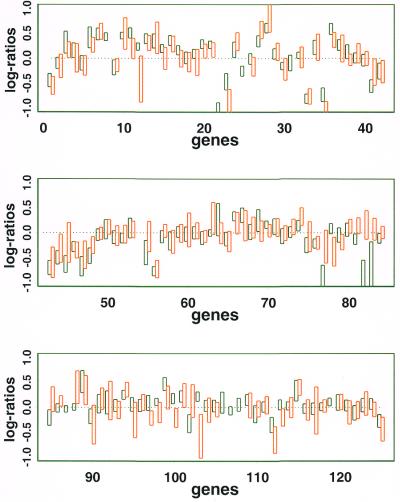
Figure 7 shows the 95% posterior interval of θg (i.e. intervals containing 95% of the probability in the posterior distribution of θg) on common genes (Table 3) in both the 125 and 4129 gene projects. We note that genes with stronger agreement of normalized log ratios across the two replicated comparative experiments have shorter intervals, as expected. The 4129 gene project generally has larger intervals than the 125 gene project, perhaps because the former is single spotted and lacks a quality filtering step.
Figure 7.
The orange and green rectangles show the 95% posterior interval for the underlying expression level θg (see text) for the 125 and 4129 gene projects (green, 125 gene project; orange, 4129 gene project). Rectangles of gene 54 (aceA) are below –1.0 and do not appear on the graph.
Table 3. Gene numbers and gene names in the 125 gene project.
| 1 | fbp | 26 | lpdA | 51 | fruR | 76 | aspC | 101 | aroH |
| 2 | pfkB | 27 | aceE | 52 | crp | 77 | aspA | 102 | aroG |
| 3 | pfkA | 28 | pdhR | 53 | cyaA | 78 | cysK | 103 | aroF |
| 4 | pgi | 29 | edd | 54 | aceA | 79 | avtA | 104 | tyrB |
| 5 | glk | 30 | tktB | 55 | frdA | 80 | rpoN | 105 | ispA |
| 6 | crr | 31 | tktA, | 56 | mdh | 81 | asnA | 106 | idi |
| 7 | ptsG | 32 | talB | 57 | htpG | 82 | metE | 107 | dxr |
| 8 | ptsHI | 33 | gltA | 58 | mopA | 83 | metB | 108 | dxs |
| 9 | pykA | 34 | adhE | 59 | grpE | 84 | leuA | 109 | ubiX |
| 10 | pykF | 35 | acs | 60 | dnaJ | 85 | lysC | 110 | ubiH |
| 11 | eno | 36 | ackA | 61 | dnaK | 86 | dapB | 111 | ubiA |
| 12 | gpmA | 37 | pta | 62 | atpA | 87 | ilvG | 112 | fadB |
| 13 | pgk | 38 | ldhA | 63 | pntA | 88 | hisG | 113 | glpD |
| 14 | gapA | 39 | fdhF | 64 | himA | 89 | trpE | 114 | glpA |
| 15 | tpiA | 40 | pflD | 65 | rpoS | 90 | ilvC | 115 | ndh |
| 16 | fba | 41 | fumB | 66 | rpoH | 91 | thrA | 116 | nuoA |
| 17 | rpe | 42 | fumC | 67 | rpoE | 92 | serA | 117 | hypB |
| 18 | rpiB | 43 | fumA | 68 | rpoD | 93 | argF | 118 | hycB |
| 19 | rpiA | 44 | sdhC | 69 | dsbA | 94 | glnA | 119 | ups |
| 20 | gnd | 45 | sucA | 70 | lepB | 95 | proB | 120 | ispB |
| 21 | zwf | 46 | icdA | 71 | secA | 96 | asnB | 121 | narH |
| 22 | pps | 47 | acnB | 72 | lon | 97 | fabA | 122 | cydA |
| 23 | pckA | 48 | acnA | 73 | glyA | 98 | pyrB | 123 | cyoA |
| 24 | ppc | 49 | arcA | 74 | gltD | 99 | purF | 124 | fdnH |
| 25 | pflB, | 50 | fnr | 75 | gdhA | 100 | aroL | 125 | poxB |
The results of the two projects show general agreement. According to the 95% posterior interval, among 119 common genes in the two projects there were 35 up-regulated and 30 down-regulated genes in the 125 gene project and 23 up-regulated and 19 down-regulated genes in the 4129 gene project. Among them there were 17 up-regulated and 17 down-regulated genes that agreed in both projects. The average sizes of the 95% intervals of normalized log ratios were 0.27 and 0.43, respectively, in the 125 and 4129 gene projects, which correspond to 0.73- to 1.4- and 0.61- to 1.6-fold changes, respectively. In the few genes that disagreed strongly between the two projects we found that most of them are grouped in certain pathways, such as metE, metB, aroL, aroG and aroF. This suggests that these strong disagreements may reflect real biological variation between the cultures used in the two different projects. We have not discussed how to account for multiple comparisons, i.e. selecting apparently differentially expressed genes from the large number of genes in the genome. Methods to account for multiple comparisons have been reviewed in Dudoit et al. (3).
Biological significance
The results of the comparative experiments in the 125 and 4129 gene projects are largely consistent with previous data (8), despite the different experimental and analytical methods used. In general the transcription pattern is consistent with the direction of metabolic flux. In acetate medium, glucose transport (ptsHI, ptsG and crr) and glycolytic genes (pfkA, fba, gapA, pgk and pykF) are down-regulated compared to the cells grown in the glucose minimal medium. Genes encoding subunits of pyruvate dehydrogenase (aceEF) are also significantly down-regulated in acetate. These results are consistent with the fact that gluose transporters, glycolysis and pyruvate dehydrogenase are not required for growth in acetate. On the other hand, genes in the TCA cycle (gltA, icdA, sdhA, sucD, mdh, fumA and fumC) and glyoxylate bypass (aceAK) are significantly up-regulated, again consistent with the need for cells grown in acetate. One of the acetate utlization genes, acs (encoding acetyl-CoA synthetase), is highly up-regulated, while the other, ackA (encoding acetate kinase), is down-regulated. This result suggests that acs may be the main acetate utilization enzyme when E.coli is growing in acetate as the sole carbon source. The gluconeogenic genes (pckA, pps and fbp) are all significantly up-regulated. The pck and fbp genes are known to be required for E.coli growth in acetate. However, a pps null mutant was shown to grow normally in acetate (17). It is possible that pps and malic enzyme form an additional pathway for gluconeogenic flux when cells are grown in acetate. Although the above metabolic genes are apparently expressed according to the needs of the cell in a particular medium, the underlying molecular mechanisms are not completely understood. Some unknown genes, such as b1725, b1518, b0598 and b1516, are highly up-regulated in acetate, while others, such as b0905, b2973, b3279 and b1903, are down-regulated. These data might give clues to the functions of these genes.
Some experimental design issues
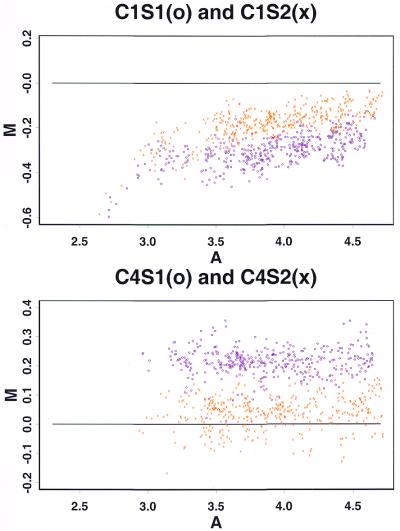
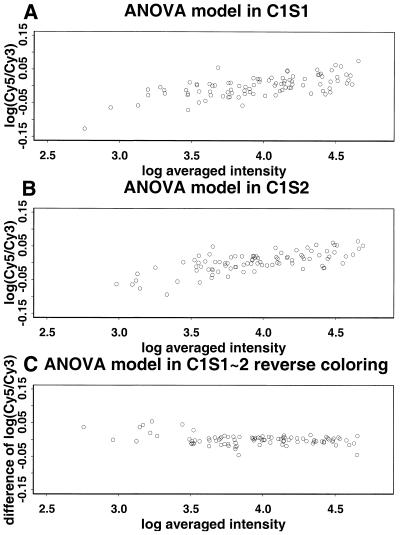
Reverse labeling and calibration experiment. In a reverse labeling design (5,18) each of the two samples (say A and B) to be compared is divided into two aliquots and labeled with two different dyes (say Cy3 and Cy5) in separate steps. Two hybridization experiments are then performed. In the first hybridization solution sample A is labeled with Cy3 and sample B is labeled with Cy5. In the second hybridization solution the labeling is reversed. We can use our calibration experiments to assess the usefulness of reverse labeling by regarding the results of the two slides in a calibration experiment, say C1S1 and C1S2, as arising from the two hybridizations of a reverse labeling experiment. This is valid since in this case all four labeling reactions were performed on aliquots derived from the same sample. Thus the calibration experiment is a special case of reverse labeling when the comparative samples A and B are identical. Figure 8A and B gives a scatter plot of the difference log(Cy5) – log(Cy3) versus the average [log(Cy3) + log(Cy5)]/2. The systematic trends that are evident in these plots are due to the inadequacy of linear normalization (see above). As a result, if an ordinary design were used then low expression genes in the Cy5-labeled comparative sample are likely to be incorrectly identified as being down-regulated. However, this problem is greatly alleviated by the reverse labeling design. The estimated gene effect log(Cy5) – log(Cy3) from these two slides (Fig. 8C) clusters tightly around the 0 line and shows no systematic trend, just as it should when the two comparative samples are identical. Thus in this example reverse labeling offers useful protection against the non-linearity of label normalization without the need to explicitly model the non-linearity. The analysis in Supplementary Material shows that such protection is not guaranteed but that partial protection can be expected under the condition that the non-linearity contributions of each gene have the same sign on both slides. Another potential benefit of reverse labeling is cancellation of the gene–label interaction (see above). A gene–label interaction can also be handled through explicit modeling, but this is not pursued here since in our experiments such a gene–label interaction is not significant compared to the other sources of variation.
Figure 8.
M–A plot for the 125 gene project. There is an increasing trend in both the first and second plots. When applying reverse labeling design the trend is largely cancelled.
Note that the reverse labeling design has the advantage of simple computation. However, when we want to perform a series of experiments, such as taking samples at different time points, the design will be more cumbersome. Performing non-linear normalization and explicit modeling of the gene–label interaction is a useful alternative.
Multiple spotting versus multiple slides. Multiple spots and multiple slides are replications to help us assess variations due to spots and slides. Since normalization is slide dependent, multiple slides information cannot be used to assess experimental quality before normalization. Thus the normalization procedure itself is vulnerable to contamination by poor quality spots. On the other hand, multiple spots within the same slide provide useful information for filtering out contaminated spots, poor quality genes or problematical slides in each experiment (see above). We also tried to apply a similar quality filtering procedure to normalized log ratios in singly spotted replicate arrays. This is less effective because the between-slide variation is typically much larger than between-spot variation, thus reducing the power for detection of outliers. In practical microarray applications it may be desirable to monitor as many genes as possible at the beginning and singly spotted arrays are more effective at this stage. However, after narrowing down the number of target genes one may be interested in using a custom array to investigate these genes further. The use of multiple spotting should be considered in the design of these arrays.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
The work of M.K.O., L.R. and J.C.L. is supported by NSF grant BES-9911718 and NIST grant 70NANBOH0064. The work of G.C.T. and W.H.W. is supported by NIH grant 1R01HG02340-01 and NSF grant DBI-9904701.
References
- 1.Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nat. Genet., 21 (suppl. 1), 33–37. [DOI] [PubMed] [Google Scholar]
- 2.Chen Y., Dougherty,E.R. and Bittner,M.L. (1997) Ratio-based decisions and the quantitative analysis of cDNA microarray images. J. Biomed. Optics, 2, 364–374. [DOI] [PubMed] [Google Scholar]
- 3.Dudoit Y., Yang,Y.H., Callow,M.J. and Speed,T.P. (2000) Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Technical Report 578, Department of Statistics, UC Berkeley, CA.
- 4.Kerr M.K. and Churchill,G.A. (2000) Experimental design for gene expression microarrays. Biostatistics, in press. [DOI] [PubMed] [Google Scholar]
- 5.Kerr M.K., Martin,M. and Churchill,G.A. (2000) Analysis of variance for gene expression microarray. J. Comput. Biol., 7, 819–837. [DOI] [PubMed] [Google Scholar]
- 6.Lee M.T., Kuo,F.C., Whitmore,G.A. and Sklar,J. (2000) Importance of replication in microarray gene expression studies: statistical methods and evidence from repetitive cDNA hybridizations. Proc. Natl Acad. Sci. USA, 18, 9834–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newton M.A., Kendziorski,C.M., Richmond,C.S., Blattner,F.R. and Tsui,K.W. (2001) On differential variability of expression ratios: improving statistical inference about gene expression changes from microarray data. J. Comput. Biol., 8, 37–52. [DOI] [PubMed] [Google Scholar]
- 8.Oh M.K. and Liao,J.C. (2000) Gene expression profiling by DNA microarrays and metabolic fluxes in Escherichia coli. Biotechnol. Prog., 16, 268–276. [DOI] [PubMed] [Google Scholar]
- 9.Oh M.K. and Liao,J.C. (2000) DNA microarray detection of metabolic responses of protein overproduction in Escherichia coli. Metab. Eng., 2, 201–209. [DOI] [PubMed] [Google Scholar]
- 10.Blattner F.R., Plunkett,G., Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F., Gregor,J., Davis,N.W., Kirkpatrick,H.A., Goeden,M.A., Rose,D.J., Mau,B. and Shao,Y. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 11.Eisen M.B. and Brown,P.O. (1999) DNA arrays for analysis of gene expression. Methods Enzymol., 303, 179–205. [DOI] [PubMed] [Google Scholar]
- 12.Miller J.H. (1992) A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 13.Venables W.N. and Ripley,B.D. (1998) Modern Applied Statistics with S-PLUS, 2nd Edn. Springer, New York, NY.
- 14.Schadt E.E., Li,C., Su,C. and Wong,W.H. (2001) Analyzing high-density oligonucleotide gene expression array data. J. Cell. Biochem., 80, 192–202. [PubMed] [Google Scholar]
- 15.Schadt E.E., Li,C., Ellis,B. and Wong,W.H. (2001) Feature extraction and normalization algorithm for high-density oligonucleotide gene expression array data. Preprints 303, Department of Statistics, UCLA, Los Angeles, CA. [DOI] [PubMed]
- 16.Gilks W.R., Richardson,S. and Speigelhalter,D.J. (1995) Markov Chain Monte Carlo in Practice. Chapman and Hall, London.
- 17.Brice C.B. and Kornberg,H.L. (1967) Location of a gene specifying phosphopyruvate synthase activity on the genome of Escherichia coli, K12. Proc. R. Soc. Lond. Ser. B Biol. Sci., 68, 281–292. [DOI] [PubMed] [Google Scholar]
- 18.Marton M.J., DeRisi,J.L., Bennett,H.A., Iyer,V.R., Meyer,M.R., Roberts,C.J., Stoughton,R., Burchard,J., Slade,D., Dai,H., Bassett,D.E., Hartwell,.L.H.,Jr, Brown,P.O. and Friend,S.H. (1998) Drug validation and identification of secondary drug target effects using DNA microarrays. Nat. Med., 4, 1293–1301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.