Abstract
As the rate of sequencing increases, greater throughput is demanded from read aligners. The full-text minute index is often used to make alignment very fast and memory-efficient, but the approach is ill-suited to finding longer, gapped alignments. Bowtie 2 combines the strengths of the full-text minute index with the flexibility and speed of hardware-accelerated dynamic programming algorithms to achieve a combination of high speed, sensitivity and accuracy.
Aligning sequencing reads to a reference genome is the first step in many comparative genomics pipelines, including pipelines for variant calling1, isoform quantitation2 and differential gene expression3. In many cases, the alignment step is the slowest. This is because for each read the aligner must solve a difficult computational problem: determining the read's likely point of origin with respect to a reference genome4.
Many aligners use a genome index to rapidly narrow the list of candidate alignment locations. The full-text minute index5 is a fast and memory-efficient index that has been used in recent aligners6–10. Index-assisted aligners work by searching for all ways of mutating the read string into a string that occurs in the reference, subject to an alignment policy limiting the number of differences. Although this search space is large, many portions of it can be skipped (‘pruned’) without loss of sensitivity. In practice, pruning strategies such as double indexing6 and bidirectional Burrows-Wheeler transform (BWT)7 facilitate very efficient ungapped alignment of short reads.
Index-aided alignment can be quite inefficient, however, when alignments are permitted to contain gaps. Alignment gaps can result either from sequencing errors or from true insertions and deletions. Ungapped aligners such as Bowtie will usually fail to align reads spanning gaps and will therefore miss evidence for these events. Gaps greatly increase the size of the search space and reduce the effectiveness of pruning, thereby substantially slowing aligners built solely on index-assisted alignment. Bowtie 2 extends the full-text minute index–based approach of Bowtie to permit gapped alignment by dividing the algorithm broadly into two stages: an initial, ungapped seed-finding stage that benefits from the speed and memory efficiency of the full-text minute index and a gapped extension stage that uses dynamic programming and benefits from the efficiency of single-instruction multiple-data (SIMD) parallel processing available on modern processors. The combination of full-text minute index–assisted seed alignment and SIMD-accelerated dynamic programming achieves an effective combination of speed, sensitivity and accuracy across a range of read lengths and sequencing technologies.
For each read, Bowtie 2 proceeds in four steps (Supplementary Note and Supplementary Fig. 1). In step 1, Bowtie 2 extracts ‘seed’ substrings from the read and its reverse complement. In step 2, the extracted substrings are aligned to the reference in an ungapped fashion assisted by the full-text minute index. In step 3, seed alignments are prioritized, and their positions in the reference genome are calculated from the index. In step 4, seeds are extended into full alignments by performing SIMD-accelerated dynamic programming.
To assess how Bowtie 2 performs on real data, we compared Bowtie 2 to three other full-text minute index–based read aligners: Burrows-Wheeler Aligner (BWA)8, BWA's Smith-Waterman alignment (BWA-SW)9 and short oligonucleotide alignment program 2 (SOAP2)10 as well as to Bowtie6. In all experiments, the reference we used was the GRCh37 major build of the human genome, including sex chromosomes, mitochondrial genome and ‘non-chromosomal’ sequences. We obtained 100-by-100 nucleotide (nt) paired-end HiSeq (2000) reads from a human resequencing study11 and extracted a random subset of 2 million pairs.
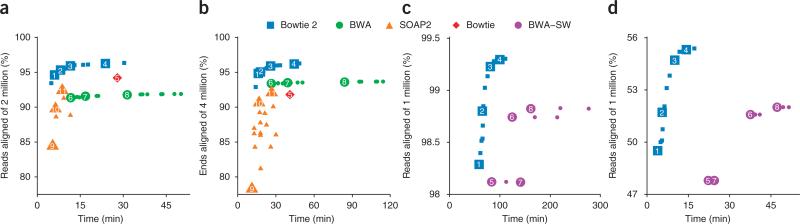
We first used BWA, SOAP2, Bowtie 2 and Bowtie to align one end (labeled ‘1’) from the subset in an unpaired fashion. To illustrate parameter tradeoffs, we ran three of the tools with a wide variety of parameter settings (Fig. 1a and Supplementary Table 1). Note that SOAP2 and Bowtie do not permit gapped alignment of unpaired reads. The Bowtie 2 default mode is faster than all BWA modes we tried and more than 2.5 times faster than the BWA default mode. All Bowtie 2 modes aligned a greater number of reads than either BWA (Supplementary Table 2) or SOAP2. The peak memory footprint of Bowtie 2 (3.24 gigabytes) was between that of BWA (2.39 gigabytes) and SOAP2 (5.34 gigabytes).
Figure 1.
Alignment comparison using HiSeq 2000, 454 and Ion Torrent reads. (a–d) Bowtie 2, BWA, SOAP2 and Bowtie were used to align two million 100 nt × 100 nt paired-end HiSeq 2000 reads from a resequencing study11. Shown are results for unpaired alignment of end 1 (a), paired-end alignment (b), Bowtie 2 and BWA-SW alignment of 1 million 454 reads from the 1000 Genomes Project Pilot12 (c), and Bowtie 2 and BWA-SW to align one million Ion Torrent reads from the G. Moore resequencing project13 (d). Plotted is the percentage of reads for which at least one alignment was found. Each numbered point is data obtained using command-line parameters shown in Supplementary Table 1.
We then aligned reads in a paired-end fashion, using a variety of alignment parameters (Fig. 1b and Supplementary Table 1). Bowtie is at a disadvantage in this scenario because it only searches for ungapped, concordant paired-end alignments. We found that the Bowtie 2 default mode was faster than all BWA modes we tried and more than 3 times faster than the BWA default mode. All Bowtie 2 modes aligned a greater number of reads than either BWA (Supplementary Table 2) or SOAP2. The peak memory footprint of Bowtie 2 (3.26 gigabytes) was similar to that of BWA (3.20 gigabytes) and smaller than that of SOAP2 (5.34 gigabytes).
To assess Bowtie 2 performance on longer reads, we obtained Roche 454 reads from the 1000 Genomes Project Pilot12 and Ion Torrent reads from the G. Moore genome resequencing project13. We extracted a random subset of 1 million reads from each and aligned them with BWA-SW and Bowtie 2. We did not align with Bowtie, BWA or SOAP2 because those tools are designed for shorter reads. We configured Bowtie 2 to perform local alignment similar to BWA-SW, and we ran both tools with various parameter settings. For both the 454 and Ion Torrent data (Fig. 1c,d and Supplementary Table 1), the Bowtie 2 default local-alignment mode was faster and aligned more reads (Supplementary Table 2) than any of the BWA-SW modes, with a smaller peak memory footprint (3.39 gigabytes for Bowtie 2 and 3.66 gigabytes for BWA-SW).
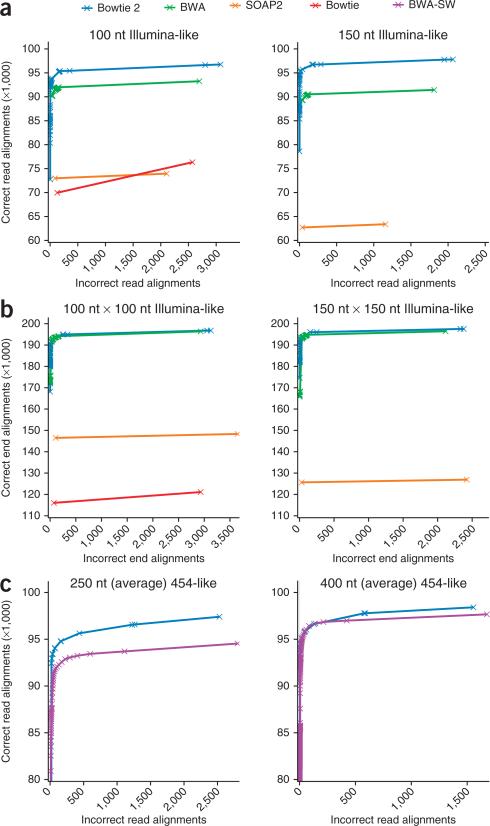
To assess the accuracy and sensitivity of Bowtie 2, we used simulated reads for which we knew the correct alignment. Using Mason (http://www.seqan.de/projects/mason.html), we simulated sets of 100,000 Illumina-like single reads 100 nt long and 150 nt long from the human genome or the same number of paired-end reads, and ran Bowtie 2, BWA and SOAP2 on each dataset. We also ran Bowtie on the 100 nt and 100 nt × 100 nt datasets. For each aligner and each dataset, we recorded the number of correct and incorrect alignments stratified by mapping quality, defined as –10 log10(p), where p is the aligner's estimate of the probability that the read was aligned incorrectly. We then calculated the cumulative number of correct and incorrect alignments from high to low mapping quality. We considered an alignment correct only if the leftmost position was within 50 nt of the position assigned by the simulator on the same strand.
We plotted cumulative correct alignments against cumulative incorrect alignments for each dataset and aligner (Fig. 2 and Supplementary Table 3). In all cases, Bowtie 2 and BWA reported more correct alignments than SOAP2 and Bowtie. For the unpaired reads, the plots indicate that Bowtie 2 gave more correct and fewer incorrect alignments than BWA over a range of mapping quality cutoffs. For paired-end reads, the difference was smaller. Note that in its paired-end mode, BWA performed local alignment to recover one of the two ends of the paired-end read in some situations, which Bowtie 2 did not. In the 150-nt dataset comparison, for instance, BWA trimmed 2,991 reads in this way. In Supplementary Results and Supplementary Figure 2 we show results for additional read lengths.
Figure 2.
Sensitivity and accuracy of alignment using simulated reads. (a–c) Cumulative number of correct and incorrect alignments from high to low mapping quality for simulated Illumina-like unpaired 100 nt and 150 nt datasets (a), for simulated Illumina-like paired-end 100 nt × 100 nt and 150 nt × 150 nt datasets (b), and for simulated 454-like datasets with average read lengths 250 nt and 400 nt (c) using indicated aligners.
We also used the Mason simulator to generate two collections of 100,000 454-like reads with average lengths of 250 nt and 400 nt. We ran Bowtie 2 and BWA-SW on these datasets (Fig. 2 and Supplementary Table 3). Bowtie 2 generally outperformed BWA-SW, especially for the 250-nt reads. BWA-SW also trimmed more reads; for the 250-nt data, for example, it trimmed 53,486 reads compared to 51,051 reads by Bowtie 2.
Full-text minute index–assisted search is an increasingly common approach for aligning sequencing reads. Extending this method to perform sensitive gapped alignment without incurring serious computational penalties is a major technical challenge. We found that Bowtie 2, a method that combines the advantages of the full-text minute index and SIMD dynamic programming, achieved very fast and memory-efficient gapped alignment of sequencing reads. Bowtie 2 improved on the previous Bowtie method in terms of speed and fraction of reads aligned (Supplementary Results, Supplementary Figs. 3, 4 and Supplementary Tables 4, 5) and was substantially faster than non–full-text minute index–based approaches while aligning a comparable fraction of reads (Supplementary Results and Supplementary Table 6). Robustness to edits, especially gaps, will continue to be a crucial concern as errors typically manifest as gaps in emerging single-molecule sequencing technologies. The speed, sensitivity, accuracy, quality-value awareness and ability to align in both local and end-to-end modes make Bowtie 2 particularly apt for current and future sequencing workloads. Bowtie 2 is free, open-source software available as Supplementary Software and at http://bowtie-bio.sourceforge.net/bowtie2/index.shtml.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturemethods/.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by US National Institutes of Health grants R01-HG006102 and R01-GM083873 to S.L.S. and an Amazon Web Services in Education Research grant to B.L. in July 2011.
ONLINE METHODS
Real data comparisons
We indexed the reference genome with each tool's default indexing parameters. Executable files for Bowtie 2 v2.0.0-beta4, Bowtie v0.12.7 and BWA 0.5.9-r16 were obtained via standard build procedures with default arguments. For SOAP2 v2.21, Linux executable files were downloaded from the tool website. ‘Running time’ was measured from initial invocation of the aligner to completion of SOAP-format or SAM-format14 output. ‘Reads/ends aligned’ was measured as the number of reads or ends for which the tool found at least one alignment regardless of mapping quality or alignment score. ‘Peak virtual memory usage’ was measured using the Linux ‘top’ utility.
For BWA, separate invocations of the software were required for aligning each end and for processing intermediate alignment results into a final SAM file. ‘Running time’ and ‘Peak virtual memory usage’ were measured across all invocations of the software. These experiments used a single Intel Xeon X5550 Nehalem 2.66 GHz processor of a high-memory extra large instance (m2.xlarge) rented the Amazon Web Services Elastic Compute Cloud (EC2) service (http://aws.amazon.com/ec2/). The instance had 17.1 gigabytes of physical memory and ran Red Hat Enterprise Linux Server release 6.1. Note that Bowtie, Bowtie 2 and BWA can align reads to the human genome on a desktop computer with 4 GB of RAM.
Unpaired HiSeq 2000 comparison
The reads used were European Nucleotide Archive accession ERR037900. To illustrate parameter tradeoffs, Bowtie 2, SOAP2 and BWA were run with a variety of parameters. In the case of Bowtie 2, we varied parameters controlling seed length (-L), spacing between seeds (-i), the number of consecutive dynamic programming attempts that can fail before giving up on a mate or read (-D) and the number of times Bowtie 2 will ‘re-seed’ for reads with repetitive seed strings (-R) (Supplementary Note). In the case of BWA, we varied parameters controlling the seed length (-l), seed differences permitted (-k) and gap opens permitted overall (-o). In the case of SOAP2, we varied parameters controlling seed length (-l) and mismatches permitted overall (-v). Note that SOAP2 does not permit gapped alignment of unpaired reads. Bowtie 2 was run in ‘end-to-end’ alignment mode, meaning that it attempted to align the entire read without omitting characters at either extreme. BWA and SOAP2 behave similarly. Bowtie was run with parameters ‘-l 28 -n 2 -e 250 -M 1–best’.
Paired HiSeq 2000 comparison
The reads used are accession ERR037900. To illustrate parameter tradeoffs, each tool was run with a variety of parameters. All the same variations were used as for the unpaired comparison. In addition, though, we varied the permitted gap size (-g) for SOAP2. For all pairedend runs, SOAP2 was run with options ‘-m 250 -x 500’, to enforce minimum and maximum fragments lengths of 250 nt and 500 nt, respectively. For Bowtie 2 and BWA, the maximum fragment length was left at its default value of 500 nt. For Bowtie, the maximum fragment length was set to 500 nt (-X 500). Bowtie 2 was run in ‘end-to-end’ alignment mode, meaning that it attempted to align the entire read without omitting characters at either extreme. BWA and SOAP2 behaved similarly, with the caveat that BWA sometimes uses local alignment when searching for one of the two ends in a paired-end read. Bowtie was run with parameters ‘-l 28 -n 2 -e 250 -M 1–best’.
454 and Ion Torrent comparisons
The 454 reads used were Sequence Read Archive accession SRR003161 and the Ion Torrent reads are European Nucleotide Archive accession ERR039480. The minimum, average and maximum read lengths for the 454 reads used were 15 nt, 355 nt and 631 nt, respectively. The minimum, average and maximum read lengths for the Ion Torrent reads were 4 nt, 191 nt and 2,716 nt, respectively.
Bowtie 2 was run in ‘local’ mode, meaning that some nucleotides at either extreme of the read could be omitted (that is, ‘soft trimmed’ or ‘soft clipped’) as determined by a Smith-Waterman–like scoring scheme. BWA-SW behaved similarly by default. Note that it is possible to adjust score thresholds in a way that aligns many more reads but trims many more bases from their extremes. To ensure that results were comparable, Bowtie 2 was run with the ‘–bwa-sw-like’ option, which caused Bowtie 2 to match BWA-SW's default score and threshold configuration (as determined by BWA-SW's -a, -b, -q, -r, -T and -c options) as closely as possible.
To illustrate parameter tradeoffs, both tools were run with a variety of parameters. In the case of Bowtie 2, we varied the same parameters as were varied in the HiSeq 2000 data comparisons. In the case of BWA-SW, we varied parameters controlling the ‘Z-best heuristic’ (-z), and a filter to remove repetitive candidate seed alignments (-s). Note that unlike BWA-SW, Bowtie 2 does not seek chimeric alignments, though the method can be extended to support this; for example, high-scoring extensions of seeds that do not to span the entire read (that is, partial alignments) could be saved and then matched with other partial alignments to form chimeric alignments when no full-length alignments were found. Support for chimeric alignment is future work.
Simulation studies
Mason 0.1 was used to simulate reads from the GRCh37 major build of the human genome, including sex chromosomes, mitochondrial genome and ‘nonchromosomal’ sequences. For the unpaired Illumina-like datasets, Mason was run in ‘Illumina’ mode with options ‘-hn 2 -sq’. For the paired-end Illumina-like datasets, Mason was run in ‘Illumina’ mode with options ‘-hn 2 -sq -mp -ll 375 -le 100’. For the 454-like datasets, Mason was run in ‘454’ mode with options ‘-hn 2 -sq -k 0.3 -bm 0.4 -bs 0.2’. For Illumina-like reads, the read length was set with the ‘-n’ option and for 454-like reads, the average read length was set with the ‘-nm’ option.
Footnotes
AUTHOR CONTRIBUTIONS
B.L. wrote the software and conducted the experiments. B.L. and S.L.S. wrote the manuscript.
Note: Supplementary information is available on the Nature Methods website.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.McKenna A, et al. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trapnell C, et al. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langmead B, Hansen KD, Leek JT. Genome Biol. 2010;11:R83. doi: 10.1186/gb-2010-11-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Homer N. Brief. Bioinform. 2010;11:473–483. doi: 10.1093/bib/bbq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferragina P, Manzini G. Proc. 41st Annual Symposium on Foundations of Computer Science. IEEE Comput. Soc.; 2000. pp. 390–398. [Google Scholar]
- 6.Langmead B, Trapnell C, Pop M, Salzberg SL. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lam T, et al. IEEE International Conference on Bioinformatics and Biomedicine. 2009:31–36. [Google Scholar]
- 8.Li H, Durbin R. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, Durbin R. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li R, et al. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 11.Ajay SS, Parker SC, Ozel Abaan H, Fuentes Fajardo KV, Margulies EH. Genome Res. 2010;21:1498–1505. doi: 10.1101/gr.123638.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.1000 Genomes Project Consortium Nature. 2010;467:1061–1073. [Google Scholar]
- 13.Rothberg JM, et al. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- 14.Li H, et al. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.