Abstract
Drosophila brain has emerged as a powerful model system for the investigation of genes being related to neurological pathologies. To map the proteomic landscape of fly brain, in a high-resolution scale, we herein employed a nano liquid chromatography-tandem mass spectrometry technology, and high-content catalogues of 7,663 unique peptides and 2,335 single proteins were generated. Protein-data processing, through UniProt, DAVID, KEGG and PANTHER bioinformatics subroutines, led to fly brain-protein classification, according to sub-cellular topology, molecular function, implication in signaling and contribution to neuronal diseases. Given the importance of Ubiquitin Proteasome System (UPS) in neuropathologies and by using the almost completely reassembled UPS, we genetically targeted genes encoding components of the ubiquitination-dependent protein-degradation machinery. This analysis showed that driving RNAi toward proteasome components and regulators, using the GAL4-elav.L driver, resulted in changes to longevity and climbing-activity patterns during aging. Our proteomic map is expected to advance the existing knowledge regarding brain biology in animal species of major translational-research value and economical interest.
Similar content being viewed by others
Introduction
Drosophila nervous system has emerged as an effective and powerful biological model for the investigation of genes involved in human neurological pathologies, including Alzheimer’s, Amyotrophic Lateral Sclerosis (ALS), Huntington’s and Parkinson’s diseases1. Most of them are characterized by age-dependent deterioration in movement coordination and anatomical disruption in specific brain regions2,3,4,5. Drosophila and human are known to present a high genetic conservation pattern, with approximately 75% of all identified human disorder-related genes having fly homologues1,6,7,8,9. Compared to the roughly 86 billion neurons of the human brain10, the entire Drosophila adult central nervous system of 150,000 neurons11,12 provides a manageable number of cells, with a great diversity of distinguishable neuronal types, and, thus, an alternative research system for identification of the genetic and neurobiological basis of a wide array of human diseases13,14. In the same context, flies typically contain a single gene ortholog, in contrast to the multiple gene paralogs found in mammals. Most importantly, Drosophila offers a powerful genetic toolbox containing transgenic methods that enable the genetic perturbation of defined neuron sets15. Another advantage is the collection of approximately 7,000 transgenic fly lines which encompass (in a variety of intersecting patterns) all neurons of the brain11,16. Furthermore, a complete, at synaptic-resolution, electron microscopy dataset of adult fly whole-brain17, behavioral models for habituation18, and a growing number of antibodies all critically contribute to the knowledge advancement of brain biology and science.
Several studies have previously shown that the physiological operation of Ubiquitin Proteasome System (UPS), one of the two major protein degradation machineries, is essential for the proper formation of neuronal networks, and for the synaptic development and plasticity4,19,20,21. Aberrations in UPS are associated, either as primary causes or secondary consequences, with development of a number of neurological pathologies, such as ALS, Alzheimer’s, Huntington’s and Parkinson’s diseases, mainly due to the accumulation of aggregation-prone neurotoxic proteins1,3,4,22,23,24.
UPS is indispensable for the maintenance of protein homeostasis, since more than 80% of all cellular proteins are degraded through its proteolytic actions. Thus, proteasomes are the second most abundant protein complexes in cells, constituting up to 5% of the total protein content25,26,27,28. Elimination of ubiquitinated short-lived, misfolded and damaged proteins is mainly performed by the 26S proteasome. For their recognition and proteolysis by proteasome (26S), target proteins are reversibly tagged with a poly-ubiquitin chain via the sequential actions of E1 (ubiquitin-activating), E2 (ubiquitin-conjugating) and E3 (ubiquitin-ligase) enzymes29,30. The reverse process of ubiquitin (Ub) removal from Ub-bound proteins is accomplished by a superfamily of more than 100 deubiquitinating enzymes (DUBs)27,31. 26S proteasome is composed of a 20S proteolytic complex (core particle/CP), being capped by one or two 19S regulatory complexes (regulatory particle/RP)28,32. 20S-CP comprises 14 different α- and β-type subunits, stacked in 4 heptameric rings being arranged in an α1–7, β1–7, β1–7 and α1–7 architecture. The β1, β2 and β5 subunits possess caspase-, trypsin- and chymotrypsin-like activities, hydrolyzing proteins after acidic, basic and hydrophobic amino acid residues, respectively. Two outer α-rings confer attachment sites for the 19S-RP and form a molecular gate through which proteins enter the catalytic site33,34. 19S-RP carries 6 Rpt (Regulatory particle of triple-ATPase) and 13 Rpn (Regulatory particle of non-ATPase) subunits, and is responsible for recognition, binding, deubiquitination, unfolding and translocation of poly-ubiquitinated proteins to 20S-CP, in an ATP-dependent manner26,27,33. It (19S-RP) can be further divided into base and lid sub-complexes. Base, which is associated with 20S proteasome, is structured by the ATPases Rpt1–6, the scaffolding proteins Rpn1 and 2, and the ubiquitin receptors Rpn10 and 13. Lid, which recognizes, binds and deubiquitinates substrate proteins, is assembled by the Rpn3, 5–9, 11 (a metalloprotease with DUB activity), 12 and 15 subunits27,33,35.
Ιn the present study, through employment of a nano Liquid Chromatography - tandem Mass Spectrometry (nLC-MS/MS) proteomics approach, we have identified a total of 2,335 individual proteins in the single-transgenic elav.L-GAL4/+ female adult (2–3-day-old) Drosophila brain. This high-content and -reliability proteome database, and its subsequent bioinformatics processing, will certainly advance the existing knowledge on brain physiology and neurodegeneration pathology, and will also provide a powerful and versatile tool for future proteome comparisons among species of strong translational-research and economical interest. Moreover, and since the contribution of specific UPS components and regulators to normal brain function remains largely unknown, we have also examined the in vivo effects of gene-specific UPS disruptions in Drosophila neuronal tissues, after engagement of the GAL4/UAS binary genetic system suitably coupled to the RNAi-based technology, and use of lifespan and climbing activity profiles as the biological system readouts. Our study revealed that genetic targeting of UPS leads to the development of component/regulator-specific neuropathology.
Materials and Methods
Drosophila melanogaster strain stocks and culturing conditions
The Drosophila melanogaster transgenic fly strains w[*]; P{w[+mC] = GAL4-elav.L}3 (BL: 8760) and w[1118]; y[1] w[*]; w[*]; P{w[+mC] = UAS-UBP2.D}2/CyO (BL: 9907) were obtained from Bloomington Drosophila Stock Center (NIH P40OD018537) (IN, USA). The D. melanogaster transgenic fly strains UAS-Rpn2_RNAi (VDRC ID: 44135), UAS-alpha5_RNAi (VDRC ID: 16105), UAS-dbeta5_RNAi (VDRC ID: 38659), UAS-beta6_RNAi (VDRC ID: 34801) and UAS-UbcD1_RNAi (VDRC ID: 26011) were provided by Vienna Drosophila Resource Center (Vienna, Austria)36. Fly stocks were maintained at 25 °C, on a 12 h light/dark cycle, and fed on standard diet (6.4% rice flour, 5% tomato paste, 3.2% sugar, 0.8% yeast, 0.8% agar, 0.4% ethanol and 0.4% propionic acid).
High-resolution proteοmics: peptide generation - nLC-MS/MS
Total-protein extracts were prepared from 100 D. melanogaster whole-brains, having been isolated, via manual dissection, from single-transgenic elav.L-GAL4/+ young adult (2–3-day-old) female flies. Protein extraction and processing were carried out as previously described37. Generated peptides were analyzed using an LTQ Orbitrap Elite instrument (Thermo Scientific, IL, USA), with the mass spectrometer being coupled to a Dionex Ultimate 3000 HPLC system. The extracted ion chromatogram was further processed using the Proteome Discoverer software (Thermo Scientific, IL, USA) and the Sequest search engine. The database chosen for protein identification searches was the D. melanogaster reference proteome, directly downloaded from UniProt 2.16 resource, without further modifications. Identification criteria included a precursor-mass tolerance of 10 ppm and fragment-mass tolerance of 0.05 Da. Trypsin was selected as the cleavage enzyme, with a maximum of “0” missed-cleavage parameter. A false-discovery rate-threshold of 0.5% ensured the reliability of protein identification procedure.
Longevity measurement
Populations of 20–25 flies (males and females in separate vials) were thoroughly analyzed in terms of their respective longevities. Survival curves were generated by daily counting the number of deceased flies. The results of each viability experiment consisted of at least 100 flies, from three different fly crosses, were statistically analyzed. All viability experiments were performed at the same time for control and RNAi-downregulated strains.
Intriguingly, ubiquitous activation of RNAi machinery, through GeneSwitch system, has been previously reported to result in RNAi sequence-independent side-effects on Drosophila lifespan under aging38. However, RNAi expression restricted to certain tissues may not be detrimental to lifespan38. In accordance, we have recently shown, through engagement of the elav.L-GAL4/UAS and RNAi genetic platforms, that neuronal cell-specific targeting of CCS (copper chaperone for SOD1) gene affected climbing activity of Drosophila female flies during aging but not their longevity39, therefore suggesting that -at least- in this system the activation of RNAi machinery alone cannot affect aging.
Climbing activity
Climbing activity (negative geotaxis assay) is a powerful in vivo indicator for the reliable evaluation of locomotor performance and, thus, neuromuscular integrity in Drosophila40,41. Climbing assay was carried out as previously described42. Briefly, 25–30 flies (males and females were being tested separately), from each experimental group, were being placed every 10 days in an empty graduated 100 ml (clean) cylinder, with a line drawn at the 66 ml (2/3) mark. Next, flies were gently tapped to the bottom of the cylinder, in order to start climbing (against gravity) all together. The number of flies that reached above the 66 ml mark, after a 20 sec time-period, was recorded. The assay was repeated 10 times for each group, allowing for 1 min rest-period in between 2 successive trials. Results were, then, converted to percentages, and the average pass rate per genotype and time point was computed. For each examined genotype, at least 5 different groups, from independent genetic crosses, per time-point were analyzed. Comparisons between control and RNAi-targeted fly-groups were carried out at the same time.
Statistical analysis
Statistical analysis was performed using the Statistical Package for Social Sciences (IBM SPSS v23.0 for Windows IBM Corp., NY, USA). For climbing assays, the results were presented graphically as an average pass rate per genotype/time-point with sample standard deviation (±SSD) value. Differences between compared genotypes were evaluated using the independent t-test analysis. All data from lifespan experiments were analyzed with the Kaplan-Meier survival test, using log rank and Breslow test statistics. Significance was accepted at p < 0.05 (*) and p < 0.01 (**).
Bioinformatics subroutines
The obtained UniProt43 protein accession numbers were processed for annotation through the DAVID bioinformatics resource (versions 6.7 and 6.8)44,45, the KEGG pathway maps46,47,48 and the PANTHER bioinformatics platform49,50.
Results and Discussion
High-resolution mapping of Drosophila melanogaster brain proteome: organelle compartmentalization and functional dissection of the mapped proteins
The elav.L-GAL4 is a strong neuronal cell-specific driver51,52,53, routinely used for gene-function analysis studies in Drosophila nervous system. Protein extracts from 100 manually dissected brains, derived from elav.L-GAL4 heterozygote D. melanogaster female flies (elav.L-GAL4/+) of 2 to 3 days old, were processed through a high-resolution nLC-MS/MS proteomics approach, and 7,663 unique peptides and 2,335 single proteins were identified (Supplementary Table 1). Out of this large collection of fly-brain proteins, 777 were classified to reside in the cell membranes (including external and internal ones), 538 in the nucleus (including nucleolus), 120 in the extracellular region and 1,124 in the cytoplasm (Fig. 1A). Furthermore, regarding cytoplasmic proteins, a number of organelle-related ones were recognized in the mitochondrion (n = 287), vesicle (n = 91), endoplasmic reticulum (n = 83), ribosome (n = 66), Golgi apparatus (n = 63), peroxisome (n = 27) and lysosome (n = 19) (Fig. 1A).
Gene Ontology-based annotation of single-transgenic elav.L-GAL4/+ Drosophila brain proteome. (A) Sub-cellular-topology classification of 2,335 fly-brain proteins having been obtained and identified via engagement of a high-resolution nLC-MS/MS proteomics technology. (B) Fly-brain proteome categorization in “Biological Processes” and “Molecular Functions”. (C) Major “Signaling Pathways” having been assembled in the fly-brain setting. Due to functional overlaps, a number of proteins are categorized in more than one group. (A,B) DAVID v.6.8 - Gene Ontology subroutine. (C) DAVID v.6.7 - PANTHER.
Next, we dissected the herein catalogued D. melanogaster brain-proteome contents into a plethora of general biological processes and specific molecular functions (Fig. 1B). The great majority of proteins seem to be implicated in metabolic processes (n = 1,045), cell communication (n = 493), gene expression (n = 449), hydrolase activity (n = 378), nucleic acid binding (n = 375), nucleotide binding (n = 350), cell cycle (n = 237), vesicle-mediated transport (n = 225) and transporter activity (n = 204) (Fig. 1B).
In an effort to reconstruct the fundamental pathways that critically control the physiology of neuronal cells, we analyzed Drosophila brain proteome via the PANTHER-pathway classification system being utilized through DAVID 6.7 bioinformatics resource (Fig. 1C). Proteins that belong to major signaling pathways and processes, such as axon guidance (n = 12) and angiogenesis (n = 25), which are crucial in neuronal-wiring mechanisms during brain development, were recognized. Pathways numerically enriched with brain proteins embrace the Wnt (n = 35), the integrin (n = 31), the FGF (n = 21), the EGF (n = 20) and the heterotrimeric G-protein pathway (n = 29). Interestingly, dopamine pathway, which besides the synthesis of neurotransmitter dopamine is also involved in functions such as learning, reward and motivation, is presented with 16 proteins (Fig. 1C).
The employment of liquid chromatography coupled with tandem-mass spectrometry has been previously employed to uncover the Drosophila head proteome of wild type flies54. In that study, a total number of 4,812 proteins were detected in an enriched membrane fraction of fly heads. More recently, Kuznetsova and collaborators analyzed the Drosophila brain proteome in a mixed population of equal male and female wild type flies of different ages, and identified 4,005 proteins55. Since the protein dataset of Kuznetsova and collaborators55 has been obtained from fly brains, albeit from diverse proteomic backgrounds, we, next, proceeded to brain proteome comparison, using UniProt accession and Flybase ID numbers, revealing approximately 1,727 shared proteins (Fig. S1). Without excluding technical differences of the employed protocols, the 608 unique proteins of our study may indicate an effect of GAL4 activity on the regulation of their expression.
Next, and since over 50% of fly genes show sequence homologies to human genes and approximately 75% of all known human-disease genes are believed to have functional fly homologues1,3,7,8,9,56, the identification in Drosophila brain proteome of proteins related to human pathologies was expected. Specifically, Drosophila brain protein-homologues of human ones associated with Parkinson’s disease (n = 26), Huntington’s disease (n = 26), Alzheimer’s disease (n = 20) and chemokine/cytokine-mediated inflammation (n = 33) were recognized (Fig. 2). These results highlight the prospect of utilizing Drosophila brain as a model system for the in vivo analysis of conserved pathways related to those being defective in human neurodegenerative diseases.
UPS molecular reconstruction in Drosophila brain
Development of a number of neurodegenerative diseases is linked to failures in UPS, which result in the accumulation of neurotoxic proteins (reviewed in1,3,4,22,23,24). Deregulation of UPS affects the length and number of axons and dendrites, while it also leads to deficient synaptic function that is believed to be an early effect of neurodegenerative diseases, rendering UPS an important pharmacological target4,20,57,58,59,60,61,62. Thereby, we focused on proteins related to UPS components and regulators. A total of 27 proteins belonging to the proteasome complex were identified in our proteomic database, 13 of which were presented in the 20S core and the remaining 14 in the 19S regulatory complex (9 to base and 5 to lid sub-complex) (Fig. 3A). Additionally, 15 proteins being associated with the ubiquitin tagging of substrate-targets destined for proteasomal degradation were also recognized. Among them, 9 proteins were linked to cullin-RING E3 ubiquitin ligases (Fig. 3A), the largest superfamily of E3 ubiquitin ligases. Notably, molecular reconstruction of the 26S Drosophila brain proteasome through KEGG-pathway maps resulted in a rather complete protein model of both 20S and 19S proteasome complexes (Fig. 3B).
Reconstruction of UPS in Drosophila brain-proteome environment. (A) Categorization of fly-brain proteins strongly involved in UPS structure, function and regulation, following their characterization through engagement of the Gene Ontology subroutine of DAVID v.6.8. (B) Molecular reassembly of Drosophila 26S proteasome, through utilization of the KEGG-pathway database46,47,48 (permission ref: 200112). Orange-colored boxes: proteins of the present study identified in the fly-brain proteome. Green-colored boxes: proteins that were absent from our study (for either technical or biological reasons). White boxes: proteins that are completely missing from Drosophila melanogaster proteome.
Targeted disruption of 26S proteasome components in Drosophila neuronal tissues results in severe lifespan reduction and kinetic pathology
Based on the importance of UPS integrity to neuropathologies, we, next, investigated the effects of proteasome impairment in Drosophila neuronal cells during aging, with the use of GAL4/UAS genetic platform. GAL4/UAS, the most widely used system in Drosophila for achieving ectopic gene expression, allows the selective activation of any cloned gene or RNAi in a wide variety of tissue- and cell- specific patterns. Briefly, in one Drosophila line the GAL4 transcriptional activator of the yeast is introduced into its genome under the control of a cell/tissue-specific endogenous promoter, while in another line the target transgene or RNAi is cloned downstream of a UAS sequence. In the progeny of a cross between these lines, the target gene/RNAi of interest is expressed in the same cell/tissue-specific pattern as the GAL4 activator63,64,65. Hence, we genetically targeted critical 26S-proteasome subunits, using the neuronal-specific elav.L-GAL4 driver. Specifically, we examined representative subunits from both 19S-RP and 20S-CP, one of which (β5) carries proteolytic activity.
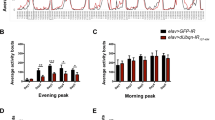
According to previous reports, Rpn2 serves as scaffolding subunit of the 19S proteasome-base ring and together with Rpn1 comprise the two largest proteasome subunits of RP, which are both necessary for the coordinate functions of substrate shuttles, ubiquitin receptors and deubiquitinating enzymes66,67,68. After downregulating the Rpn2 subunit, specifically in neuronal cells, using the RNAi-based technology, a reduction of approximately 20 days in the viability of both male and female flies was observed at their 50% survival rate, as compared to control populations (Fig. 4A,B). Likewise, their climbing activity, especially in male flies, was also negatively affected, following an age-dependent declining pattern (Fig. 5A,B). Altogether, Rpn2 seems to represent an essential proteasome subunit controlling neuronal proteostasis. These findings are strongly supported by a previous report from our laboratory, describing the development of distinct, dysmorphic phenotypes in Drosophila eye and wing, in response to RNAi-mediated downregulation of Rpn1 or Rpn2 proteasome subunit69.
Neuronal cell-specific targeting of UPS key subunits in Drosophila. Lifespan profiles of Drosophila male (left panels) and female (right panels) transgenic fly populations, being characterized by RNAi-mediated targeting of the (A,B) Rpn2 19S-RP subunit (elav.L > Rpn2_RNAi), (C,D) α5 20S-CP subunit (elav.L > α5_RNAi), (E,F) β5 20S-CP catalytic subunit (elav.L > β5_RNAi), (G,H) β6 20S-CP subunit (elav.L > β6_RNAi) and (I,J) UbcD1 E2 enzyme (elav.L > UbcD1_RNAi), specifically in neuronal tissues. **p < 0.001.
Genetic targeting of UPS in neuronal tissues causes climbing pathology in Drosophila. Climbing-activity patterns of male (left panels) and female (right panels) transgenic flies, carrying downregulated protein contents of the (A,B) Rpn2 19S-RP subunit (elav.L > Rpn2_RNAi), (C,D) α5 20S-CP subunit (elav.L > α5_RNAi), (E,F) β5 20S-CP catalytic subunit (elav.L > β5_RNAi), (G,H) β6 20S-CP subunit (elav.L > β6_RNAi) and (I,J) UbcD1 E2 enzyme (elav.L > UbcD1_RNAi), specifically in neuronal tissues. *p < 0.05.
We, next, proceeded to investigate the role of outer α-rings of 20S-CP in normal nervous system. These are comprised of 7 different α subunits (α1–7), which provide the binding sites for 19S-RP and form the molecular gate responsible for regulating substrate entrance into the inner catalytic cavity of β-rings28,70. Flies with reduced levels of α5 proteasome subunit, specifically in neuronal cells, of both sexes, managed to survive only for few days (Fig. 4C,D), while they also presented dramatically diminished climbing activities (Fig. 5C,D), thus underscoring the indispensable contribution of α5 subunit to nervous-system physiological development and function in Drosophila. Similarly to Rpn2, the tissue-specific downregulated expression of α5 subunit in Drosophila has also been previously associated with developmentally malformed eyes and wings69. Interestingly, the critical role of α5 subunit in proteasomal activity has been demonstrated in yeast, with α5 mutations (such as the substitution of an α-pocket lysine residue) giving rise to proteasome holoenzymes that carry immature β subunits and reduced peptidase activities71. Strikingly, mutations in α5 subunit of Saccharomyces cerevisiae have proved to notably affect its lifespan and revealed that the 20S-CP gate opening is directly controlled by the α5 subunit72.
The inner β-rings are composed of the inactive (unable to hydrolyze) β3, β4, β6, and β7 subunits, and the active β1, β2 and β5 subunits, which contain proteolytic sites of different specificities. The β5 subunit with a chymotrypsin-like activity hydrolyzes proteins after hydrophobic amino acid residues and carries the major proteasome-related proteolytic activity33,73. We herein show that male and female flies being depleted of either β5 (Fig. 4E,F) or β6 (Fig. 4G,H) proteasome subunit, specifically in neuronal tissues, are presented with a life expectancy of only few days. Furthermore, they are characterized by total climbing (kinetic) deficiency, given their (both sexes) inability to pass the specified cylinder mark during the respective climbing assay (Fig. 5E–H). These observations are in line with previous ones, describing flies that bear a dominant-negative mutation in the β6 gene, which die as undifferentiated pupae with failures in head eversion at a restrictive temperature74. This mutation has been also reported to alter cell-fate determination in the adult Drosophila sense-organ lineage75. It has been previously proposed that a deficient β6-proteasome subunit may disrupt proteasomal activity by altering the β2-β6 interfacing regions75,76. Importantly, previous studies have associated β5 and β6 proteasome-subunit deregulation with disrupted Drosophila eye and wing development69,77,78. In accordance to our findings, data from transgenic mice have shown that reduction in β5-associated chymotrypsin-like activity leads to multiple early-aging phenotypes and shortened lifespan79. On the contrary, overexpression of β5 subunit can improve proteostasis during aging and can increase longevity in Drosophila80. Ιt seems that certain thresholds of 26S-proteasome activities are required for Drosophila nervous-system physiological development and function.
RNAi-mediated downregulation of the E2 ubiquitin-conjugating enzyme UbCD1 specifically in Drosophila neuronal tissues compromises longevity and climbing activity
Ubiquitination of substrates destined for proteasomal degradation depends on the sequential actions of ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s) and ubiquitin-ligases (E3s), which mediate the final transfer of ubiquitin to the selected substrates81. E2s contain a highly conserved 150–200 amino acid ubiquitin-conjugating catalytic-core domain and are involved in the transfer of ubiquitin or ubiquitin-like proteins to downstream conjugation targets. E2s are responsible for the selection of correct modifier, the determination of length, type and topology of attached ubiquitin chain, and the processivity of chain-assembly reaction, thereby defining the fate assignment of the modified protein73,81,82,83,84. It has recently emerged that E2s, besides their typical role in protein degradation, are also involved in processes controlling protein function, sorting and localization, while their aberrant regulation has been associated with many disease pathways81,85. Among them, UbcD1, or effete, encodes a highly conserved E2 enzyme, with approximately 80% sequence identity to the yeast Ubc4/Ubc5 homologue, which mediates the selective protein-ubiquitination and degradation processes86. UbcD1 has been also associated with proper telomere behavior during cell division87, Hedgehog signaling by controlling Ci stability88, female germline stem-cell maintenance89, and regulation of apoptosis through its involvement in the poly-ubiquitination and degradation of DIAP190,91. UbcD1 has been directly associated with Drosophila SCF complex of proteasome-dependent proteolysis92,93,94 and, as it is illustrated in Fig. 3A, the cullin-RING E3 ligase (SCF) complex can be readily detected in Drosophila nervous system.
Therefore, in an in vivo attempt to prove the importance of neuronal-specific UbcD1 targeting to the lifespan and climbing activity of D. melanogaster during aging, we genetically downregulated its expression using the elav.L-GAL4 and UbcD1_RNAi transgenic flies. UbcD1-targeted male (Fig. 4I) and female (Fig. 4J) flies were presented with a maximum life expectancy of only 20 days, while their vast majority were dying within the first 10 days. Notably, both male (Fig. 5I) and female (Fig. 5J) fly populations appeared with completely impaired climbing activities, since none of the tested flies proved capable to climb over the specified cylinder mark. Similarly, another study has reported that among 16 genes encoding E2 enzymes, only the RNAi-mediated UbcD1 targeting resulted in neuroblast overgrowth94.
Protein deubiquitination deregulation in neuronal cells during Drosophila aging
Ubiquitination, the post-translational modification of proteins by ubiquitin, can be reversed by deubiquitinating enzymes (DUBs), a specific family of proteases that catalyze the removal of ubiquitin (or ubiquitin-like) molecules from target substrates, modulating protein stability and signaling31,33. DUBs are divided into 6 structurally distinct subfamilies based on the architecture of their catalytic domains and mechanisms of action31,33,95,96. In the human genome, approximately 100 DUBs are encoded, while in the fly there have been identified around 45-family members31,95,97,98. DUBs serve as key regulators of several intracellular processes, such as NFκB activation (USP11 and 15), c-Myc stability (USP28), p53 stability (USP2a, 7 and 10), DNA-damage response (USP1) and apoptosis (USP7 and 28)27,31,95,96,99. Aberrant expression patterns and mutant forms of DUBs have been implicated in pathogenesis of several human diseases, including cancer and neurological pathologies31,95,96,98, and, as such, DUBs have emerged to represent attractive drug targets for pharmacological intervention strategies27,29,99.
Ubiquitous suppression of most DUBs in Drosophila has been associated with severe deregulation of fly development, motility and longevity98. Hence, in an effort to in vivo examine the significance of deubiquitination process in neuronal tissues during Drosophila aging, we herein generated transgenic flies carrying downregulated expression of the dUBP64 protease (a deubiquitinating enzyme). Usp47, which is the human ortholog of the fly dUBP64 deubiquitinating enzyme, has been found to interact with the E3 ubiquitin ligase SCF100 and to play a key role in the control of axonal growth during neuronal development101, while its depletion from different cell lines has been associated with decreased cell survival100. In Drosophila, dUBP64 functions as a modifier of position-effect variegation102 and controller of cell-fate decision during eye development, by regulating the transcriptional repressor tramtrack103.
In contrast to male transgenic flies (Fig. 6A), female populations, at their 50% survival rate, were presented with a reduction of approximately 40 days in their viability, as compared to control populations (Fig. 6B). Moreover, both male and female dUBP64-targeted flies were characterized by similar to controls climbing activities (Fig. 7A,B). Similar fly-sex-specific differential longevity profiles have been also reported in response to pathogen infection and altered mitochondrial dynamics104,105. Furthermore, to investigate the effects of DUB overexpression in lifespan and climbing activity, transgenic flies overexpressing the -yeast- UBP2 protease (a deubiquitinating enzyme), specifically in neuronal tissues, were generated through utilization of the elav.L-GAL4 driver. Both male (Fig. 6C) and female (Fig. 6D) transgenic flies were presented with significantly reduced viabilities and climbing activities (Fig. 7C,D), as compared to control populations. It seems that UBP2 overexpression critically compromises Drosophila longevity and climbing activity, and that neuronal cells are unable to counterbalance, or neutralize, the proteotoxic stress being induced by UBP2 abundance. Ubiquitination pathway is intrinsically required for the regulation of synaptic growth and function in Drosophila, with the -yeast- UBP2 deubiquitinating enzyme overexpression specifically in nervous system leading to synaptic overgrowth and defect in neurotransmitter release106. Overexpression of UBP2 has been also associated with synaptic-elimination delay in postsynaptic muscles107. Axon and dendrite pruning are fundamental for development of proper circuitry in Drosophila nervous system, and UBP2 elevated contents can cause blockage of both axon and dendrite pruning, and degeneration108,109,110.
Genetic deregulation of ubiquitinated protein-load homeostasis in Drosophila neuronal tissues. Fly-longevity profiles of male (left panels) and female (right panels) transgenic flies, bearing either downregulated protein contents (via employment of RNAi technology) of the (A,B) dUBP64 deubiquitinating enzyme (elav.L > dUBP64_RNAi), or elevated (overexpressed) protein levels of the (C,D) yeast UBP2 deubiquitinating enzyme (elav.L > UBP2_yeast), specifically in neuronal tissues. **p < 0.001.
Effects of neuronal-specific deregulation of protein deubiquitination process in Drosophila climbing. Climbing-activity patterns of male (left panels) and female (right panels) transgenic flies, carrying either downregulated protein contents of the (A,B) dUBP64 deubiquitinating enzyme (elav.L > dUBP64_RNAi), or increased protein levels of the (C,D) yeast UBP2 deubiquitinating enzyme (elav.L > UBP2_yeast), specifically in neuronal tissues. *p < 0.05.
Overall, we herein mapped, in a high-resolution scale, the brain proteomic landscape of the single-transgenic elav.L-GAL4/+ D. melanogaster fly strain. This, together with the availability of fly-mutant (genetic) lines able to model some aspects of human neuropathology (reviewed in1,7,56,111), may enable the screen of a large number of drugs, which are expected to be favorably exploited for further elucidating disease mechanisms, and identifying novel druggable targets and disease therapies. Indeed, researchers have recently used genetically modified Drosophila flies, as a personalized biological platform, for discovering the best (optimum) drug therapy being applied to a patient carrying treatment-resistant colorectal cancer112. Conclusively, fine tuning of ubiquitinated-protein homeostasis in neuronal cells has herein proved essential for animal’s well-being during aging.
References
Bier, E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6, 9–23, https://doi.org/10.1038/nrg1503 (2005).
Lloyd, T. E. & Taylor, J. P. Flightless flies: Drosophila models of neuromuscular disease. Ann. N. Y. Acad. Sci. 1184, e1–20 (2010).
Bonini, N. M. & Fortini, M. E. Human neurodegenerative disease modeling using Drosophila. Annu. Rev. Neurosci. 26, 627–656, https://doi.org/10.1146/annurev.neuro.26.041002.131425 (2003).
Jaiswal, M., Sandoval, H., Zhang, K., Bayat, V. & Bellen, H. J. Probing mechanisms that underlie human neurodegenerative diseases in Drosophila. Annu. Rev. Genet. 46, 371–396, https://doi.org/10.1146/annurev-genet-110711-155456 (2012).
Mutsuddi, M. & Nambu, J. R. Neural disease: Drosophila degenerates for a good cause. Curr. Biol. 8, R809–811 (1998).
Oortveld, M. A. et al. Human intellectual disability genes form conserved functional modules in Drosophila. PLoS Genet. 9, e1003911, https://doi.org/10.1371/journal.pgen.1003911 (2013).
Pandey, U. B. & Nichols, C. D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharmacol. Rev. 63, 411–436, https://doi.org/10.1124/pr.110.003293 (2011).
Rubin, G. M. et al. Comparative genomics of the eukaryotes. Science 287, 2204–2215, https://doi.org/10.1126/science.287.5461.2204 (2000).
Reiter, L. T., Potocki, L., Chien, S., Gribskov, M. & Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 11, 1114–1125, https://doi.org/10.1101/gr.169101 (2001).
Azevedo, F. A. et al. Equal numbers of neuronal and nonneuronal cells make the human brain an isometrically scaled-up primate brain. J. Comp. Neurol. 513, 532–541, https://doi.org/10.1002/cne.21974 (2009).
Jenett, A. et al. A GAL4-driver line resource for Drosophila neurobiology. Cell Rep. 2, 991–1001, https://doi.org/10.1016/j.celrep.2012.09.011 (2012).
Taniguchi, H. & Moore, A. W. Chromatin regulators in neurodevelopment and disease: Analysis of fly neural circuits provides insights: Networks of chromatin regulators and transcription factors underlie Drosophila neurogenesis and cognitive defects in intellectual disability and neuropsychiatric disorder models. Bioessays 36, 872–883, https://doi.org/10.1002/bies.201400087 (2014).
Zwarts, L., Clements, J. & Callaerts, P. In The Making and Un-Making of Neuronal Circuits in Drosophila Neuromethods (ed Bassem A. Hassan) Ch. Chapter 1, 3–48 (Humana Press, 2012).
O’Kane, C. J. Drosophila as a model organism for the study of neuropsychiatric disorders. Curr. Top. Behav. Neurosci. 7, 37–60, https://doi.org/10.1007/7854_2010_110 (2011).
Venken, K. J., Simpson, J. H. & Bellen, H. J. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron 72, 202–230, https://doi.org/10.1016/j.neuron.2011.09.021 (2011).
Pfeiffer, B. D. et al. Tools for neuroanatomy and neurogenetics in Drosophila. Proc. Natl Acad. Sci. USA 105, 9715–9720, https://doi.org/10.1073/pnas.0803697105 (2008).
Zheng, Z. et al. A Complete Electron Microscopy Volume of the Brain of Adult Drosophila melanogaster. Cell 174, 730–743 e722, https://doi.org/10.1016/j.cell.2018.06.019 (2018).
Engel, J. E. & Wu, C. F. Neurogenetic approaches to habituation and dishabituation in Drosophila. Neurobiol. Learn. Mem. 92, 166–175, https://doi.org/10.1016/j.nlm.2008.08.003 (2009).
Ramirez, J. et al. Proteomic Analysis of the Ubiquitin Landscape in the Drosophila Embryonic Nervous System and the Adult Photoreceptor Cells. PLoS One 10, e0139083, https://doi.org/10.1371/journal.pone.0139083 (2015).
Ding, M. & Shen, K. The role of the ubiquitin proteasome system in synapse remodeling and neurodegenerative diseases. Bioessays 30, 1075–1083, https://doi.org/10.1002/bies.20843 (2008).
Patrick, G. N. Synapse formation and plasticity: recent insights from the perspective of the ubiquitin proteasome system. Curr. Opin. Neurobiol. 16, 90–94, https://doi.org/10.1016/j.conb.2006.01.007 (2006).
Ciechanover, A. & Brundin, P. The ubiquitin proteasome system in neurodegenerative diseases: sometimes the chicken, sometimes the egg. Neuron 40, 427–446 (2003).
Zheng, Q. et al. Dysregulation of Ubiquitin-Proteasome System in Neurodegenerative Diseases. Front. Aging Neurosci. 8, 303, https://doi.org/10.3389/fnagi.2016.00303 (2016).
Giasson, B. I. & Lee, V. M. Are ubiquitination pathways central to Parkinson’s disease? Cell 114, 1–8, https://doi.org/10.1016/s0092-8674(03)00509-9 (2003).
Marguerat, S. et al. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683, https://doi.org/10.1016/j.cell.2012.09.019 (2012).
Collins, G. A. & Goldberg, A. L. The Logic of the 26S Proteasome. Cell 169, 792–806, https://doi.org/10.1016/j.cell.2017.04.023 (2017).
D’Arcy, P., Wang, X. & Linder, S. Deubiquitinase inhibition as a cancer therapeutic strategy. Pharmacol. Ther. 147, 32–54, https://doi.org/10.1016/j.pharmthera.2014.11.002 (2015).
Gu, Z. C. & Enenkel, C. Proteasome assembly. Cell Mol. Life Sci. 71, 4729–4745, https://doi.org/10.1007/s00018-014-1699-8 (2014).
Mansour, M. A. Ubiquitination: Friend and foe in cancer. Int. J. Biochem. Cell Biol. 101, 80–93, https://doi.org/10.1016/j.biocel.2018.06.001 (2018).
Pickart, C. M. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70, 503–533, https://doi.org/10.1146/annurev.biochem.70.1.503 (2001).
Komander, D., Clague, M. J. & Urbe, S. Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563, https://doi.org/10.1038/nrm2731 (2009).
Raynes, R., Pomatto, L. C. & Davies, K. J. Degradation of oxidized proteins by the proteasome: Distinguishing between the 20S, 26S, and immunoproteasome proteolytic pathways. Mol. Asp. Med. 50, 41–55, https://doi.org/10.1016/j.mam.2016.05.001 (2016).
Tanaka, K. The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 12–36 (2009).
Grigoreva, T. A., Tribulovich, V. G., Garabadzhiu, A. V., Melino, G. & Barlev, N. A. The 26S proteasome is a multifaceted target for anti-cancer therapies. Oncotarget 6, 24733–24749, https://doi.org/10.18632/oncotarget.4619 (2015).
Lander, G. C. et al. Complete subunit architecture of the proteasome regulatory particle. Nature 482, 186–191, https://doi.org/10.1038/nature10774 (2012).
Dietzl, G. et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156, https://doi.org/10.1038/nature05954 (2007).
Velentzas, A. D. et al. Global Proteomic Profiling of Drosophila Ovary: A High-resolution, Unbiased, Accurate and Multifaceted Analysis. Cancer genomics Proteom. 12, 369–384 (2015).
Alic, N. et al. Detrimental effects of RNAi: a cautionary note on its use in Drosophila ageing studies. PLoS One 7, e45367, https://doi.org/10.1371/journal.pone.0045367 (2012).
Theotoki, E. I. et al. Targeting of copper-trafficking chaperones causes gene-specific systemic pathology in Drosophila melanogaster: prospective expansion of mutational landscapes that regulate tumor resistance to cisplatin. Biol Open 8, https://doi.org/10.1242/bio.046961 (2019).
Southwood, C. M., Garbern, J., Jiang, W. & Gow, A. The unfolded protein response modulates disease severity in Pelizaeus-Merzbacher disease. Neuron 36, 585–596, https://doi.org/10.1016/s0896-6273(02)01045-0 (2002).
Vernace, V. A., Arnaud, L., Schmidt-Glenewinkel, T. & Figueiredo-Pereira, M. E. Aging perturbs 26S proteasome assembly in Drosophila melanogaster. FASEB J. 21, 2672–2682, https://doi.org/10.1096/fj.06-6751com (2007).
Velentzas, P. D. et al. Detrimental effects of proteasome inhibition activity in Drosophila melanogaster: implication of ER stress, autophagy, and apoptosis. Cell Biol. Toxicol. 29, 13–37, https://doi.org/10.1007/s10565-012-9235-9 (2013).
UniProt: a hub for protein information. Nucleic Acids Res 43, D204-212, https://doi.org/10.1093/nar/gku989 (2015).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13, https://doi.org/10.1093/nar/gkn923 (2009).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57, https://doi.org/10.1038/nprot.2008.211 (2009).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein science: a Publ. Protein Soc. 28, 1947–1951, https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K. & Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47, D590–D595, https://doi.org/10.1093/nar/gky962 (2019).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30, https://doi.org/10.1093/nar/28.1.27 (2000).
Mi, H., Muruganujan, A., Casagrande, J. T. & Thomas, P. D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 8, 1551–1566, https://doi.org/10.1038/nprot.2013.092 (2013).
Mi, H., Muruganujan, A. & Thomas, P. D. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 41, D377–386, https://doi.org/10.1093/nar/gks1118 (2013).
Robinow, S. & White, K. Characterization and spatial distribution of the ELAV protein during Drosophila melanogaster development. J. Neurobiol. 22, 443–461, https://doi.org/10.1002/neu.480220503 (1991).
Kaya-Copur, A. & Schnorrer, F. A Guide to Genome-Wide In Vivo RNAi Applications in Drosophila. Methods Mol. Biol. 1478, 117–143, https://doi.org/10.1007/978-1-4939-6371-3_6 (2016).
Luo, L., Liao, Y. J., Jan, L. Y. & Jan, Y. N. Distinct morphogenetic functions of similar small GTPases: Drosophila Drac1 is involved in axonal outgrowth and myoblast fusion. Genes. Dev. 8, 1787–1802, https://doi.org/10.1101/gad.8.15.1787 (1994).
Aradska, J. et al. Gel-free mass spectrometry analysis of Drosophila melanogaster heads. Proteomics 15, 3356–3360, https://doi.org/10.1002/pmic.201500092 (2015).
Kuznetsova, K. G. et al. Brain Proteome of Drosophila melanogaster Is Enriched with Nuclear Proteins. Biochemistry 84, 71–78, https://doi.org/10.1134/S0006297919010097 (2019).
Yamamoto, S. et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell 159, 200–214, https://doi.org/10.1016/j.cell.2014.09.002 (2014).
Hamilton, A. M. & Zito, K. Breaking it down: the ubiquitin proteasome system in neuronal morphogenesis. Neural Plast. 2013, 196848, https://doi.org/10.1155/2013/196848 (2013).
Asano, S. et al. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347, 439–442, https://doi.org/10.1126/science.1261197 (2015).
Zheng, C., Geetha, T. & Babu, J. R. Failure of ubiquitin proteasome system: risk for neurodegenerative diseases. Neuro-degenerative Dis. 14, 161–175, https://doi.org/10.1159/000367694 (2014).
Aleong, R., Aumont, N., Dea, D. & Poirier, J. Non-steroidal anti-inflammatory drugs mediate increased in vitro glial expression of apolipoprotein E protein. Eur. J. Neurosci. 18, 1428–1438, https://doi.org/10.1046/j.1460-9568.2003.02869.x (2003).
Selkoe, D. J. Alzheimer’s disease is a synaptic failure. Science 298, 789–791, https://doi.org/10.1126/science.1074069 (2002).
Korhonen, L. & Lindholm, D. The ubiquitin proteasome system in synaptic and axonal degeneration: a new twist to an old cycle. J. Cell Biol. 165, 27–30, https://doi.org/10.1083/jcb.200311091 (2004).
Southall, T. D., Elliott, D. A. & Brand, A. H. The GAL4 System: A Versatile Toolkit for Gene Expression in Drosophila. CSH Protoc. 2008, pdb top49, https://doi.org/10.1101/pdb.top49 (2008).
McGuire, S. E., Roman, G. & Davis, R. L. Gene expression systems in Drosophila: a synthesis of time and space. Trends Genet. 20, 384–391, https://doi.org/10.1016/j.tig.2004.06.012 (2004).
Brand, A. H. & Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415 (1993).
Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513, https://doi.org/10.1146/annurev.biochem.78.081507.101607 (2009).
Rosenzweig, R., Bronner, V., Zhang, D., Fushman, D. & Glickman, M. H. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J. Biol. Chem. 287, 14659–14671, https://doi.org/10.1074/jbc.M111.316323 (2012).
Jiang, T. X., Zhao, M. & Qiu, X. B. Substrate receptors of proteasomes. Biol. Rev. Camb. Philos. Soc. 93, 1765–1777, https://doi.org/10.1111/brv.12419 (2018).
Velentzas, P. D. et al. Proteasome, but not autophagy, disruption results in severe eye and wing dysmorphia: a subunit- and regulator-dependent process in Drosophila. PLoS One 8, e80530, https://doi.org/10.1371/journal.pone.0080530 (2013).
Livneh, I., Cohen-Kaplan, V., Cohen-Rosenzweig, C., Avni, N. & Ciechanover, A. The life cycle of the 26S proteasome: from birth, through regulation and function, and onto its death. Cell Res. 26, 869–885, https://doi.org/10.1038/cr.2016.86 (2016).
Park, S., Kim, W., Tian, G., Gygi, S. P. & Finley, D. Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J. Biol. Chem. 286, 36652–36666, https://doi.org/10.1074/jbc.M111.285924 (2011).
Leme, J. M. M. et al. Mutations of Cys and Ser residues in the alpha5-subunit of the 20S proteasome from Saccharomyces cerevisiae affects gating and chronological lifespan. Arch. Biochem. Biophys. 666, 63–72, https://doi.org/10.1016/j.abb.2019.03.012 (2019).
Jung, T., Catalgol, B. & Grune, T. The proteasomal system. Mol. Asp. Med. 30, 191–296, https://doi.org/10.1016/j.mam.2009.04.001 (2009).
Saville, K. J. & Belote, J. M. Identification of an essential gene, l(3)73Ai, with a dominant temperature-sensitive lethal allele, encoding a Drosophila proteasome subunit. Proc. Natl Acad. Sci. USA 90, 8842–8846, https://doi.org/10.1073/pnas.90.19.8842 (1993).
Schweisguth, F. Dominant-negative mutation in the beta2 and beta6 proteasome subunit genes affect alternative cell fate decisions in the Drosophila sense organ lineage. Proc. Natl Acad. Sci. USA 96, 11382–11386, https://doi.org/10.1073/pnas.96.20.11382 (1999).
Smyth, K. A. & Belote, J. M. The dominant temperature-sensitive lethal DTS7 of Drosophila melanogaster encodes an altered 20S proteasome beta-type subunit. Genetics 151, 211–220 (1999).
Belote, J. M. & Fortier, E. Targeted expression of dominant negative proteasome mutants in Drosophila melanogaster. Genesis 34, 80–82, https://doi.org/10.1002/gene.10131 (2002).
Groll, M. et al. The catalytic sites of 20S proteasomes and their role in subunit maturation: a mutational and crystallographic study. Proc. Natl Acad. Sci. USA 96, 10976–10983, https://doi.org/10.1073/pnas.96.20.10976 (1999).
Tomaru, U. et al. Decreased proteasomal activity causes age-related phenotypes and promotes the development of metabolic abnormalities. Am. J. Pathol. 180, 963–972, https://doi.org/10.1016/j.ajpath.2011.11.012 (2012).
Nguyen, N. N. et al. Proteasome beta5 subunit overexpression improves proteostasis during aging and extends lifespan in Drosophila melanogaster. Sci. Rep. 9, 3170, https://doi.org/10.1038/s41598-019-39508-4 (2019).
Ye, Y. & Rape, M. Building ubiquitin chains: E2 enzymes at work. Nat. Rev. Mol. Cell Biol. 10, 755–764, https://doi.org/10.1038/nrm2780 (2009).
Valimberti, I., Tiberti, M., Lambrughi, M., Sarcevic, B. & Papaleo, E. E2 superfamily of ubiquitin-conjugating enzymes: constitutively active or activated through phosphorylation in the catalytic cleft. Sci. Rep. 5, 14849, https://doi.org/10.1038/srep14849 (2015).
van Wijk, S. J. & Timmers, H. T. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB J. 24, 981–993, https://doi.org/10.1096/fj.09-136259 (2010).
Stewart, M. D., Ritterhoff, T., Klevit, R. E. & Brzovic, P. S. E2 enzymes: more than just middle men. Cell Res. 26, 423–440, https://doi.org/10.1038/cr.2016.35 (2016).
Wenzel, D. M., Stoll, K. E. & Klevit, R. E. E2s: structurally economical and functionally replete. Biochem. J. 433, 31–42, https://doi.org/10.1042/BJ20100985 (2011).
Treier, M., Seufert, W. & Jentsch, S. Drosophila UbcD1 encodes a highly conserved ubiquitin-conjugating enzyme involved in selective protein degradation. EMBO J. 11, 367–372 (1992).
Cenci, G. et al. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes. Dev. 11, 863–875, https://doi.org/10.1101/gad.11.7.863 (1997).
Pan, C. et al. UbcD1 regulates Hedgehog signaling by directly modulating Ci ubiquitination and processing. EMBO Rep. 18, 1922–1934, https://doi.org/10.15252/embr.201643289 (2017).
Chen, D. et al. Effete-mediated degradation of Cyclin A is essential for the maintenance of germline stem cells in Drosophila. Development 136, 4133–4142, https://doi.org/10.1242/dev.039032 (2009).
Yoo, S. J. Grim stimulates Diap1 poly-ubiquitination by binding to UbcD1. Mol. Cell 20, 446–451 (2005).
Ryoo, H. D., Bergmann, A., Gonen, H., Ciechanover, A. & Steller, H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat. Cell Biol. 4, 432–438, https://doi.org/10.1038/ncb795 (2002).
Ohlmeyer, J. T. & Schupbach, T. Encore facilitates SCF-Ubiquitin-proteasome-dependent proteolysis during Drosophila oogenesis. Development 130, 6339–6349, https://doi.org/10.1242/dev.00855 (2003).
Bocca, S. N., Muzzopappa, M., Silberstein, S. & Wappner, P. Occurrence of a putative SCF ubiquitin ligase complex in Drosophila. Biochem. Biophys. Res. Commun. 286, 357–364, https://doi.org/10.1006/bbrc.2001.5394 (2001).
Li, S. et al. The SCFSlimb E3 ligase complex regulates asymmetric division to inhibit neuroblast overgrowth. EMBO Rep. 15, 165–174, https://doi.org/10.1002/embr.201337966 (2014).
Mevissen, T. E. T. & Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 86, 159–192, https://doi.org/10.1146/annurev-biochem-061516-044916 (2017).
Leznicki, P. & Kulathu, Y. Mechanisms of regulation and diversification of deubiquitylating enzyme function. J. Cell Sci. 130, 1997–2006, https://doi.org/10.1242/jcs.201855 (2017).
Zhang, J., Liu, M., Su, Y., Du, J. & Zhu, A. J. A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling. G3 2, 1563–1575, https://doi.org/10.1534/g3.112.003780 (2012).
Tsou, W. L. et al. Systematic analysis of the physiological importance of deubiquitinating enzymes. PLoS One 7, e43112, https://doi.org/10.1371/journal.pone.0043112 (2012).
He, M. et al. The emerging role of deubiquitinating enzymes in genomic integrity, diseases, and therapeutics. Cell Biosci. 6, 62, https://doi.org/10.1186/s13578-016-0127-1 (2016).
Peschiaroli, A., Skaar, J. R., Pagano, M. & Melino, G. The ubiquitin-specific protease USP47 is a novel beta-TRCP interactor regulating cell survival. Oncogene 29, 1384–1393, https://doi.org/10.1038/onc.2009.430 (2010).
Yang, S. W. et al. USP47 and C terminus of Hsp70-interacting protein (CHIP) antagonistically regulate katanin-p60-mediated axonal growth. J. Neurosci. 33, 12728–12738, https://doi.org/10.1523/JNEUROSCI.0698-13.2013 (2013).
Henchoz, S., De Rubertis, F., Pauli, D. & Spierer, P. The dose of a putative ubiquitin-specific protease affects position-effect variegation in Drosophila melanogaster. Mol. Cell Biol. 16, 5717–5725, https://doi.org/10.1128/mcb.16.10.5717 (1996).
Bajpe, P. K. et al. Deubiquitylating enzyme UBP64 controls cell fate through stabilization of the transcriptional repressor tramtrack. Mol. Cell Biol. 28, 1606–1615, https://doi.org/10.1128/MCB.01567-07 (2008).
Shahrestani, P. et al. Sexual dimorphism in Drosophila melanogaster survival of Beauveria bassiana infection depends on core immune signaling. Sci. Rep. 8, 12501, https://doi.org/10.1038/s41598-018-30527-1 (2018).
Tower, J. Mitochondrial maintenance failure in aging and role of sexual dimorphism. Arch. Biochem. Biophys. 576, 17–31, https://doi.org/10.1016/j.abb.2014.10.008 (2015).
DiAntonio, A. et al. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature 412, 449–452, https://doi.org/10.1038/35086595 (2001).
Liu, Z., Chen, Y., Wang, D., Wang, S. & Zhang, Y. Q. Distinct presynaptic and postsynaptic dismantling processes of Drosophila neuromuscular junctions during metamorphosis. J. Neurosci. 30, 11624–11634, https://doi.org/10.1523/JNEUROSCI.0410-10.2010 (2010).
Watts, R. J., Hoopfer, E. D. & Luo, L. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system. Neuron 38, 871–885 (2003).
Kuo, C. T., Jan, L. Y. & Jan, Y. N. Dendrite-specific remodeling of Drosophila sensory neurons requires matrix metalloproteases, ubiquitin-proteasome, and ecdysone signaling. Proc. Natl Acad. Sci. USA 102, 15230–15235, https://doi.org/10.1073/pnas.0507393102 (2005).
Tao, J. & Rolls, M. M. Dendrites have a rapid program of injury-induced degeneration that is molecularly distinct from developmental pruning. J. Neurosci. 31, 5398–5405, https://doi.org/10.1523/JNEUROSCI.3826-10.2011 (2011).
Buckingham, S. D., Esmaeili, B., Wood, M. & Sattelle, D. B. RNA interference: from model organisms towards therapy for neural and neuromuscular disorders. Hum. Mol. Genet. 13(Spec No 2), R275–288, https://doi.org/10.1093/hmg/ddh224 (2004).
Bangi, E. et al. A personalized platform identifies trametinib plus zoledronate for a patient with KRAS-mutant metastatic colorectal cancer. Sci. Adv. 5, eaav6528, https://doi.org/10.1126/sciadv.aav6528 (2019).
Acknowledgements
The authors wish to thank Bloomington Stock Center (NIH P40OD018537) (IN, USA) and Vienna Drosophila Resource Center (VDRC) (Vienna, Austria) for fly stocks. DJS devotes the present article to the memory of his beloved mother, who suddenly passed away in October 2015.
Author information
Authors and Affiliations
Contributions
A.D.V.: Methodology, Supervision, Writing - Original Draft. S.A.K.: Methodology, Validation. N.E.S.: Methodology, Validation. M.M.T.: Methodology, Validation. A.K.A.: Methodology, Validation, Data Curation. V.E.M.: Methodology, Writing - Original Draft. E.I.T.: Data Curation. A.F.G.: Data Curation. K.E.K.: Data Curation. I.S.P.: Data Curation. G.T.h.T.: Methodology, Validation, Data Curation. D.J.S.: Conceptualization, Resources, Writing - Review & Editing, Visualization, Project Administration, Funding Acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Velentzas, A.D., Katarachia, S.A., Sagioglou, N.E. et al. Proteomic mapping of Drosophila transgenic elav.L-GAL4/+ brain as a tool to illuminate neuropathology mechanisms. Sci Rep 10, 5430 (2020). https://doi.org/10.1038/s41598-020-62510-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-62510-0