Abstract
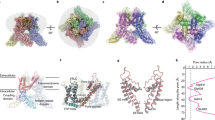
Transient receptor potential vanilloid (TRPV) cation channels are polymodal sensors involved in a variety of physiological processes. TRPV2, a member of the TRPV family, is regulated by temperature, by ligands, such as probenecid and cannabinoids, and by lipids. TRPV2 has been implicated in many biological functions, including somatosensation, osmosensation and innate immunity. Here we present the atomic model of rabbit TRPV2 in its putative desensitized state, as determined by cryo-EM at a nominal resolution of ∼4 Å. In the TRPV2 structure, the transmembrane segment 6 (S6), which is involved in gate opening, adopts a conformation different from the one observed in TRPV1. Structural comparisons of TRPV1 and TRPV2 indicate that a rotation of the ankyrin-repeat domain is coupled to pore opening via the TRP domain, and this pore opening can be modulated by rearrangements in the secondary structure of S6.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Ramsey, I.S., Delling, M. & Clapham, D.E. An introduction to TRP channels. Annu. Rev. Physiol. 68, 619–647 (2006).
Bandell, M., Macpherson, L.J. & Patapoutian, A. From chills to chilis: mechanisms for thermosensation and chemesthesis via thermoTRPs. Curr. Opin. Neurobiol. 17, 490–497 (2007).
Caterina, M.J. et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–824 (1997).
Julius, D. TRP channels and pain. Annu. Rev. Cell Dev. Biol. 29, 355–384 (2013).
Cao, E., Cordero-Morales, J.F., Liu, B., Qin, F. & Julius, D. TRPV1 channels are intrinsically heat sensitive and negatively regulated by phosphoinositide lipids. Neuron 77, 667–679 (2013).
Ufret-Vincenty, C.A. et al. Mechanism for phosphoinositide selectivity and activation of TRPV1 ion channels. J. Gen. Physiol. 145, 431–442 (2015).
Caterina, M.J., Rosen, T.A., Tominaga, M., Brake, A.J. & Julius, D. A capsaicin-receptor homologue with a high threshold for noxious heat. Nature 398, 436–441 (1999).
Perálvarez-Marín, A., Doñate-Macian, P. & Gaudet, R. What do we know about the transient receptor potential vanilloid 2 (TRPV2) ion channel? FEBS J. 280, 5471–5487 (2013).
Qin, N. et al. TRPV2 is activated by cannabidiol and mediates CGRP release in cultured rat dorsal root ganglion neurons. J. Neurosci. 28, 6231–6238 (2008).
Penna, A. et al. PI3-kinase promotes TRPV2 activity independently of channel translocation to the plasma membrane. Cell Calcium 39, 495–507 (2006).
Shibasaki, K., Murayama, N., Ono, K., Ishizaki, Y. & Tominaga, M. TRPV2 enhances axon outgrowth through its activation by membrane stretch in developing sensory and motor neurons. J. Neurosci. 30, 4601–4612 (2010).
Zanou, N. et al. Osmosensation in TRPV2 dominant negative expressing skeletal muscle fibres. J. Physiol. 593, 3849–3863 (2015).
Katanosaka, Y. et al. TRPV2 is critical for the maintenance of cardiac structure and function in mice. Nat. Commun. 5, 3932 (2014).
Link, T.M. et al. TRPV2 has a pivotal role in macrophage particle binding and phagocytosis. Nat. Immunol. 11, 232–239 (2010).
Monet, M. et al. Role of cationic channel TRPV2 in promoting prostate cancer migration and progression to androgen resistance. Cancer Res. 70, 1225–1235 (2010).
Cao, E., Liao, M., Cheng, Y. & Julius, D. TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–118 (2013).
Liao, M., Cao, E., Julius, D. & Cheng, Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–112 (2013).
Jin, X., Touhey, J. & Gaudet, R. Structure of the N-terminal ankyrin repeat domain of the TRPV2 ion channel. J. Biol. Chem. 281, 25006–25010 (2006).
McCleverty, C.J., Koesema, E., Patapoutian, A., Lesley, S.A. & Kreusch, A. Crystal structure of the human TRPV2 channel ankyrin repeat domain. Protein Sci. 15, 2201–2206 (2006).
Huynh, K.W. et al. Structural insight into the assembly of TRPV channels. Structure 22, 260–268 (2014).
Yao, J., Liu, B. & Qin, F. Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc. Natl. Acad. Sci. USA 108, 11109–11114 (2011).
Gregorio-Teruel, L. et al. The integrity of the TRP domain is pivotal for correct TRPV1 channel gating. Biophys. J. 109, 529–541 (2015).
Joseph, J., Wang, S., Lee, J., Ro, J.Y. & Chung, M.K. Carboxyl-terminal domain of transient receptor potential vanilloid 1 contains distinct segments differentially involved in capsaicin- and heat-induced desensitization. J. Biol. Chem. 288, 35690–35702 (2013).
Ufret-Vincenty, C.A., Klein, R.M., Hua, L., Angueyra, J. & Gordon, S.E. Localization of the PIP2 sensor of TRPV1 ion channels. J. Biol. Chem. 286, 9688–9698 (2011).
Brauchi, S., Orta, G., Salazar, M., Rosenmann, E. & Latorre, R. A hot-sensing cold receptor: C-terminal domain determines thermosensation in transient receptor potential channels. J. Neurosci. 26, 4835–4840 (2006).
Brauchi, S. et al. Dissection of the components for PIP2 activation and thermosensation in TRP channels. Proc. Natl. Acad. Sci. USA 104, 10246–10251 (2007).
Smart, O.S., Neduvelil, J.G., Wang, X., Wallace, B.A. & Sansom, M.S. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360, 376 (1996).
Tang, L. et al. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 505, 56–61 (2014).
Gouaux, E. & Mackinnon, R. Principles of selective ion transport in channels and pumps. Science 310, 1461–1465 (2005).
Cooley, R.B., Arp, D.J. & Karplus, P.A. Evolutionary origin of a secondary structure: π-helices as cryptic but widespread insertional variations of α-helices that enhance protein functionality. J. Mol. Biol. 404, 232–246 (2010).
Grandl, J. et al. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat. Neurosci. 13, 708–714 (2010).
Jordt, S.E., Tominaga, M. & Julius, D. Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc. Natl. Acad. Sci. USA 97, 8134–8139 (2000).
Cui, Y. et al. Selective disruption of high sensitivity heat activation but not capsaicin activation of TRPV1 channels by pore turret mutations. J. Gen. Physiol. 139, 273–283 (2012).
Ryu, S., Liu, B., Yao, J., Fu, Q. & Qin, F. Uncoupling proton activation of vanilloid receptor TRPV1. J. Neurosci. 27, 12797–12807 (2007).
Yang, F. et al. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat. Chem. Biol. 11, 518–524 (2015).
Jordt, S.E. & Julius, D. Molecular basis for species-specific sensitivity to “hot” chili peppers. Cell 108, 421–430 (2002).
Long, S.B., Tao, X., Campbell, E.B. & MacKinnon, R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature 450, 376–382 (2007).
Vieira-Pires, R.S. & Morais-Cabral, J.H. 310 helices in channels and other membrane proteins. J. Gen. Physiol. 136, 585–592 (2010).
Lishko, P.V., Procko, E., Jin, X., Phelps, C.B. & Gaudet, R. The ankyrin repeats of TRPV1 bind multiple ligands and modulate channel sensitivity. Neuron 54, 905–918 (2007).
Mercado, J., Gordon-Shaag, A., Zagotta, W.N. & Gordon, S.E. Ca2+-dependent desensitization of TRPV2 channels is mediated by hydrolysis of phosphatidylinositol 4,5-bisphosphate. J. Neurosci. 30, 13338–13347 (2010).
Suloway, C. et al. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151, 41–60 (2005).
Lander, G.C. et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166, 95–102 (2009).
Mindell, J.A. & Grigorieff, N. Accurate determination of local defocus and specimen tilt in electron microscopy. J. Struct. Biol. 142, 334–347 (2003).
Voss, N.R., Yoshioka, C.K., Radermacher, M., Potter, C.S. & Carragher, B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J. Struct. Biol. 166, 205–213 (2009).
Ludtke, S.J., Baldwin, P.R. & Chiu, W. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128, 82–97 (1999).
Ogura, T., Iwasaki, K. & Sato, C. Topology representing network enables highly accurate classification of protein images taken by cryo electron-microscope without masking. J. Struct. Biol. 143, 185–200 (2003).
Scheres, S.H. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012).
Li, X. et al. Electron counting and beam-induced motion correction enable near-atomic-resolution single-particle cryo-EM. Nat. Methods 10, 584–590 (2013).
Roseman, A.M. FindEM: a fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 145, 91–99 (2004).
Scheres, S.H. Beam-induced motion correction for sub-megadalton cryo-EM particles. eLife 3, e03665 (2014).
Heymann, J.B. & Belnap, D.M. Bsoft: image processing and molecular modeling for electron microscopy. J. Struct. Biol. 157, 3–18 (2007).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Adams, P.D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010).
Brown, A. et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr. D Biol. Crystallogr. 71, 136–153 (2015).
Fernández, I.S., Bai, X.C., Murshudov, G., Scheres, S.H. & Ramakrishnan, V. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157, 823–831 (2014).
Peier, A.M. et al. A TRP channel that senses cold stimuli and menthol. Cell 108, 705–715 (2002).
Acknowledgements
Cryo-EM data were collected at the electron microscopy facility at The Scripps Research Institute. We are indebted to J. Grandl and J. Sosa, who provided access to their calcium-imaging apparatus and guidance in calcium-imaging experiments. We thank J.-C. Ducom at The Scripps High Performance Computing facility for aiding with computational needs, S. Thomas and Z. Ouyang for initial TRPV2 biochemistry and E. Mashalidis for advice on the manuscript. This work was supported by start-up funds from the Duke University Medical Center (S.-Y.L.) and the US National Institutes of Health (R01GM100894 and DP2OD008380 to S.-Y.L., and DP2EB020402 to G.C.L.). G.C.L. is supported as a Searle Scholar and a Pew Scholar.
Author information
Authors and Affiliations
Contributions
L.Z. conducted all biochemical preparation of TRPV2 and calcium-imaging experiments under the guidance of S.-Y.L. M.A.H. conducted all electron microscopy experiments and the single-particle 3D reconstruction under the guidance of G.C.L., B.C.C. and L.Z. S.-Y.L. carried out model building and refinement. Z.L. performed electrophysiology experiments. L.Z., M.A.H., G.C.L. and S.-Y.L. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Integrated supplementary information
Supplementary Figure 1 Sequence alignment of TRPV1 and TRPV2.
Sequence numbers are based on rabbit TRPV2. The truncated loop is shown in the red box.
Supplementary Figure 2 The WT and truncated rabbit TRPV2 are activated by 2-APB.
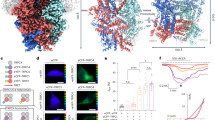
(a)-(d) Whole cell recordings of full length (a-b) and truncated rabbit TRPV2 (c-d) channels expressed in HEK293T cells. (a) and (c) Currents were evoked by ramp protocol (black squares: -120 mV; open circles:-120 mV) at the interval time of 5 seconds. The application of 1 mM 2-APB is indicated by the bar. Characters indicate basal currents, a, peak responses, b, and recovery currents, c. (b) and (d) Representative current-voltage relationship of rabbit TRPV2 at time points indicated in (a) and (c). (e) Comparison of current density of the WT (left, n=14) and the truncated rabbit TRPV2 (right, n=16) recorded at +120 mV (black bars) and -120 mV (grey bars). The data represent mean ± s.e.m. The dotted line indicates the baseline. (f-h) Increase in calcium uptake upon application of 100 μM 2-APB in cells expressing the WT rabbit TRPV2, the truncated rabbit TRPV2 and empty vector respectively. The average responses of 50-80 cells from representative experiments are shown (mean +/- s.e.m.). Traces for both transfected (GFP) and non-transfected cells (-GFP) are shown as control experiments. (i) Average fold increase in the ratio of 340/380 nm recorded from 6-8 experiments from 2 independent transfections in response to 100 μM 2-APB (mean +/- s.e.m.).
Supplementary Figure 3 EM data collection.
(a) Raw micrograph of negatively stained particles (top) and reference-free 2D class averages (middle). A low resolution negative stain reconstruction was determined (bottom) and used as an initial model for cryo-EM structure determination. (b) Representative cryo-EM micrograph of TRPV2 particles suspended in vitreous ice (top). Particles selected by automated particle selection shown on the right half of the image. Only images exhibiting Thon rings beyond 4Å (blue circle, middle) were used for image processing, as assessed by a 1D plot (bottom). (c) Raw particles extracted from the micrographs. (d) Reference-free 2D class averages showing well-ordered secondary structure in the transmembrane region of the complex.
Supplementary Figure 4 3D classification and refinement.
Initial classification of 427,360 particles into five 3D classes (I), two of which (classes 4 and 5) were used for subsequent 3D refinement. After performing the particle polishing step in RELION (II), particles were subjected to another 25 iterations of 3D classification into four classes (III). The particles contributing to the class exhibiting the highest level of structural detail was used for the final 3D refinement (IV).
Supplementary Figure 5 Radiation damage weighting and resolution assessment.
(a) Per-frame radiation damage weighting was applied by estimating the B Factor for each frame (above). The frequency-dependent weights used to generate the final stack of summed particle images is shown below. (b) Fourier Shell Correlation plots calculated from independently refined half-maps, refined using different 3D masks during refinement. The masks are shown in the upper right, and are colored to correspond to the FSC plot. (c). Final cryoEM map colored according to the local resolution, as determined by windowed FSCs of independently refined half-maps. (d) Euler distribution plot, with regions in red denoting the views containing the highest number of particles. (e) Cross-validation of the atomic model. FSC curves between the refined atomic model and the half-map 1 (solid black line), half map 2 (solid gray line), subset of polished and masked map (dotted line), and summed map (dash-dot line). The red dashed line demarcates FSC = 0.143.
Supplementary Figure 6 Structural details of the ARD and its interactions.
(a) A close-up view of the linker domain. The domain has two distinct regions: the helix-loop-helix (HLH) region and the β sheet region. CTD that covers the linker domain is omitted for clarity. (b) Close-up view of the intersubunit interactions formed by the β sheet region of the linker domain. The linker domain interacts with ankyrin repeats 3 and 4 of the neighboring subunit. Dashed line represents the missing segment of the finger 3. The distal part of the CTD contributes a β strand to the β sheet of the linker domain. (c) Superposition of the rat TRPV2 ARD (magenta, PDB 2ETA) with the ARD of the rabbit TRPV2 structure (blue). The two structures diverge around the ankyrin repeat 1 and the fingers 1-3. (d) A close-up view of the coupling domain and the TRP domain with the arginine and lysine residues displayed as sticks and marked with circles. The map covering the CTD was of lower quality compared to the rest of the molecue which prevented the building of all side-chains. Where side-chains of the Arg and Lys residues were not built, we display the backbone only. Arg682, which resides in CTD, is labeld as it is the residue corresponding to Arg721 in TRPV1 which has been identified as important for binding of PIP2.
Supplementary Figure 7 Secondary structures within the TRPV1 and TRPV2 transmembrane helices.
(a) The S6 helix in the TRPV1 channel assumes a π conformation in the middle of the helix, where the carbonyl of residue i forms a hydrogen bond with the amide of residue i+5. The dashed lines represent carbonyl-amide bonds. (b) Electron density of TRPV2 S5 and S6. The density is contoured at 1.2 sigma, with an applied B-factor of -76 Å2. (c) The S4b of TRPV2 assumes a 310 helical conformation, where the carbonyl of residue i forms a hydrogen bond with the amide of residue i+3. The dashed lines represent carbonyl-amide bonds. (d) The lower part of the TRPV1 S5, in the vicinity of the S4-S5 linker, contains a π segment. The dashed lines represent carbonyl-amide bonds. For simplicity, in all panels only one set of hydrogen bonds is shown per one turn of the helix.
Supplementary Figure 8 Coupling at the pore loop and turret and S6 and TRP domain.
(a) Side-view of the pore loop, pore helix and the turret shown in sphere representation. The interactions network between the turret and the pore helix in open TRPV1 (green) and TRPV2 (blue) are disrupted. (b) Electron density around the pore loop, pore helix and turret of TRPV2. (c) A superposition of the TRPV1 structures in open (green) and closed (red) conformations with TRPV2 (blue) shows that the HLH of TRPV2 assumes a different conformation and the S6-proximal part of the TRP domain has slipped down towards the cytosol. (d) Overlay of S5, S6 and the TRP domain of the closed (red) and open (green) TRPV1 and TRPV2 (blue). The helical connection between the S6 and the TRP domain is lost in TRPV2.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–8 and Supplementary Table 1 (PDF 3681 kb)
Structural transitions between TRPV1 closed, TRPV1 open and TRPV2
Ankyrin repeats are colored in cyan, the coupling domain is colored in orange, the TRP domain in red and the S6 in dark blue. In the closed TRPV1 state, Ile673 forms the lower gate. By contrast, in the TRPV2 structure, Met643 forms a hydrophobic seal at the helix bundle crossing. This might be caused by a π-helix to α-helix transition in S6 (indicated in green). This transition might also cause unwinding of the helical linker between the TRP domain and S6 in the TRPV2 structure. (MP4 78098 kb)
Source data
Rights and permissions
About this article
Cite this article
Zubcevic, L., Herzik, M., Chung, B. et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat Struct Mol Biol 23, 180–186 (2016). https://doi.org/10.1038/nsmb.3159
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.3159