Abstract
Protein aggregates and damaged organelles are tagged with ubiquitin chains to trigger selective autophagy. To initiate mitophagy, the ubiquitin kinase PINK1 phosphorylates ubiquitin to activate the ubiquitin ligase parkin, which builds ubiquitin chains on mitochondrial outer membrane proteins, where they act to recruit autophagy receptors. Using genome editing to knockout five autophagy receptors in HeLa cells, here we show that two receptors previously linked to xenophagy, NDP52 and optineurin, are the primary receptors for PINK1- and parkin-mediated mitophagy. PINK1 recruits NDP52 and optineurin, but not p62, to mitochondria to activate mitophagy directly, independently of parkin. Once recruited to mitochondria, NDP52 and optineurin recruit the autophagy factors ULK1, DFCP1 and WIPI1 to focal spots proximal to mitochondria, revealing a function for these autophagy receptors upstream of LC3. This supports a new model in which PINK1-generated phospho-ubiquitin serves as the autophagy signal on mitochondria, and parkin then acts to amplify this signal. This work also suggests direct and broader roles for ubiquitin phosphorylation in other autophagy pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Svenning, S. & Johansen, T. Selective autophagy. Essays Biochem. 55, 79–92 (2013)
Stolz, A., Ernst, A. & Dikic, I. Cargo recognition and trafficking in selective autophagy. Nature Cell Biol. 16, 495–501 (2014)
Narendra, D. P. et al. PINK1 is selectively stabilized on impaired mitochondria to activate parkin. PLoS Biol. 8, e1000298 (2010)
Narendra, D., Tanaka, A., Suen, D. F. & Youle, R. J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 (2008)
Kane, L. A. et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205, 143–153 (2014)
Kazlauskaite, A. et al. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460, 127–139 (2014)
Koyano, F. et al. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature 510, 162–166 (2014)
Geisler, S. et al. PINK1/Parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nature Cell Biol. 12, 119–131 (2010)
Wong, Y. C. & Holzbaur, E. L. F. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl Acad. Sci. USA 111, E4439–E4448 (2014)
Narendra, D., Kane, L. A., Hauser, D. N., Fearnley, I. M. & Youle, R. J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106 (2010)
Okatsu, K. et al. p62/SQSTM1 cooperates with Parkin for perinuclear clustering of depolarized mitochondria. Genes Cells 15, 887–900 (2010)
Lu, K., Psakhye, I. & Jentsch, S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 158, 549–563 (2014)
Wild, P. et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 333, 228–233 (2011)
Rezaie, T. et al. Adult-onset primary open-angle glaucoma caused by mutations in optineurin. Science 295, 1077–1079 (2002)
Maruyama, H. et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature 465, 223–226 (2010)
Ellinghaus, D. et al. Association between variants of PRDM1 and NDP52 and Crohn's disease, based on exome sequencing and functional studies. Gastroenterology 145, 339–347 (2013)
Mankouri, J. et al. Optineurin negatively regulates the induction of IFNβ in response to RNA virus infection. PLoS Pathog. 6, e1000778 (2010)
Thurston, T. L., Ryzhakov, G., Bloor, S., von Muhlinen, N. & Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nature Immunol. 10, 1215–1221 (2009)
Morton, S., Hesson, L., Peggie, M. & Cohen, P. Enhanced binding of TBK1 by an optineurin mutant that causes a familial form of primary open angle glaucoma. FEBS Lett. 582, 997–1002 (2008)
Larabi, A. et al. Crystal structure and mechanism of activation of TANK-binding kinase 1. Cell Rep. 3, 734–746 (2013)
Ordureau, A. et al. Quantitative proteomics reveal a feedforward mechanism for mitochondrial PARKIN translocation and ubiquitin chain synthesis. Mol. Cell 56, 360–375 (2014)
Wauer, T. et al. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 34, 307–325 (2015)
Kondapalli, C. et al. PINK1 is activated by mitochondrial membrane potential depolarization and stimulates Parkin E3 ligase activity by phosphorylating Serine 65. Open Biol. 2, 120080 (2012)
Sarraf, S. A. et al. Landscape of the PARKIN-dependent ubiquitylome in response to mitochondrial depolarization. Nature 496, 372–376 (2013)
Shiba-Fukushima, K. et al. Phosphorylation of mitochondrial polyubiquitin by PINK1 promotes Parkin mitochondrial tethering. PLoS Genet. 10, e1004861 (2014)
Okatsu, K. et al. Phosphorylated ubiquitin chain is the genuine Parkin receptor. J. Cell Biol. 209, 111–128 (2015)
Ordureau, A. et al. Defining roles of PARKIN and ubiquitin phosphorylation by PINK1 in mitochondrial quality control using a ubiquitin replacement strategy. Proc. Natl Acad. Sci. USA 112, 6637–6642 (2015)
von Muhlinen, N. et al. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell 48, 329–342 (2012)
Wild, P., McEwan, D. G. & Dikic, I. The LC3 interactome at a glance. J. Cell Sci. 127, 3–9 (2014)
Lamb, C. A., Yoshimori, T. & Tooze, S. A. The autophagosome: origins unknown, biogenesis complex. Nature Rev. Mol. Cell Biol. 14, 759–774 (2013)
Fogel, A. I. et al. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol. Cell. Biol. 33, 3675–3688 (2013)
Koyama-Honda, I., Itakura, E., Fujiwara, T. K. & Mizushima, N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 9, 1491–1499 (2013)
Kim, J., Kundu, M., Viollet, B. & Guan, K. L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nature Cell Biol. 13, 132–141 (2011)
Itakura, E., Kishi-Itakura, C., Koyama-Honda, I. & Mizushima, N. Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125, 1488–1499 (2012)
Nezich, C. L., Wang, C., Fogel, A. I. & Youle, R. J. MiT/TFE transcription factors are activated during mitophagy downstream of Parkin and Atg5. J. Cell Biol. 210, 435–450 (2015)
Santel, A. et al. Mitofusin-1 protein is a generally expressed mediator of mitochondrial fusion in mammalian cells. J. Cell Sci. 116, 2763–2774 (2003)
Huang, P. et al. Heritable gene targeting in zebrafish using customized TALENs. Nature Biotechnol. 29, 699–700 (2011)
Hasson, S. A. et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature 504, 291–295 (2013)
Miller, J. C. et al. A TALE nuclease architecture for efficient genome editing. Nature Biotechnol. 29, 143–148 (2011)
Mali, P. et al. RNA-guided human genome engineering via Cas9. Science 339, 823–826 (2013)
Lazarou, M., Jin, S. M., Kane, L. A. & Youle, R. J. Role of PINK1 binding to the TOM complex and alternate intracellular membranes in recruitment and activation of the E3 ligase Parkin. Dev. Cell 22, 320–333 (2012)
Katayama, H., Kogure, T., Mizushima, N., Yoshimori, T. & Miyawaki, A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18, 1042–1052 (2011)
Acknowledgements
We thank C. Nezich and S. Banerjee in the Youle laboratory, C. Smith and the NINDS and NHLBI Flow Cytometry Core Facilities. This work was supported by the Intramural Research Program of the NIH, NINDS and the National Health and Medical Research Council (GNT1063781).
Author information
Authors and Affiliations
Contributions
M.L., D.A.S., L.A.K. and R.J.Y. conceived the projects; M.L., D.A.S., L.A.K., S.A.S., C.W., D.P.S., A.I.F. and R.J.Y. designed experiments; M.L., D.A.S., L.A.K., S.A.S., C.W., J.L.B., D.P.S. and A.I.F. performed experiments; M.L., D.A.S., L.A.K. and R.J.Y. wrote the manuscript, and all authors contributed to editing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Extended data figures and tables
Extended Data Figure 1 Analysis of knockout cell lines and characterization of autophagy receptor translocation to damaged mitochondria.
a, The ATG5 knockout cell line was confirmed by immunoblotting. b, Representative images of mtDNA nucleoids in HeLa cells immunostained with an anti-DNA antibody (green), confirming colocalization with the mitochondrial marker TOM20 (red) (n = 3 experiments). c, Mitochondrial fractions from mCherry–parkin expressing pentaKO and wild-type cells were assessed by immunoblotting. d, mCh–parkin-expressing wild-type, pentaKO and ATG5 knockouts were treated with oligomycin/antimycin A with or without MG132. Cell lysates were assessed by immunoblotting. e, Expression levels of GFP-tagged OPTN, NDP52, p62, NBR1 and TAX1BP1 re-expressed in pentaKO cells by immunoblotting. f, Representative images of mCh–parkin-expressing pentaKOs from e immunostained for TOM20 (n = 3 experiments). g, Expression of GFP–TOLLIP in mCh–parkin pentaKOs. h, PentaKOs mCh–parkin and with or without GFP–TOLLIP expression were immunoblotted. i, Representative images of mCh–parkin pentaKOs expressing GFP–TOLLIP immunostained for TOM20 (n = 3 experiments). Scale bars, 10 μm.
Extended Data Figure 2 OPTN, NDP52 and TAX1BP1 triple knockout analysis and disease-associated mutations.
a, b, Knockout cell lines with or without mCh–parkin expression were immunoblotted (a) and COXII levels were quantified (b). c, A panel of human tissue lysates was immunoblotted. d, Expression of wild-type or mutant GFP–OPTN in mCh–parkin pentaKO cells. e, Quantification of cells in f. More than 100 cells per condition. f, Representative images of mCh–parkin pentaKO cells expressing GFP–OPTN mutants immunostained for TOM20 (n = 3). g, PentaKOs expressing mCh–parkin were rescued with wild-type or mutant GFP–OPTN, analysed by immunoblotting. See Fig. 2e for quantification of COXII. h, Expression of wild-type or mutant GFP–NDP52 in mCh–parkin pentaKO cells. i, Quantification of cells in j. More than 100 cells per condition. j, Representative images of mCh–parkin pentaKOs expressing wild-type or mutant GFP–NDP52 were immunostained for TOM20 (n = 3). k, PentaKO cells expressing mCh–parkin rescued with wild-type or mutant GFP–NDP52 were analysed by immunoblotting. See Fig. 2f for quantification of COXII. Data are mean ± s.d. from three (b and i) and two (e) independent experiments. ***P < 0.001 (one-way ANOVA). Scale bars, 10 μm.
Extended Data Figure 3 TBK1 activates OPTN in PINK1/parkin mitophagy.
a, Representative images of untreated mCh–parkin cells and merged images of treated cells as indicated immunostained for DNA. See Fig. 2a for anti-DNA and DAPI images of treated samples (n = 3). b, Cell lysates from wild-type, NDP52/OPTN DKO, OPTN knockout and NDP52 knockout cells with or without mCh–parkin expression were immunoblotted for TBK1 activation. c, Cell lysates from wild-type and PINK1 knockout cells without parkin expression were immunoblotted for TBK1 activation (S172 phosphorylation). d, Confirmation of TBK1/NDP52 DKO, TBK1/OPTN DKO and TBK1 knockout by immunoblotting. e, Knockout cell lines from d were immunostained for TOM20 (n = 3). f, Representative images of untreated mCh–parkin wild-type and knockout cells, and merged images of treated cells as indicated were immunostained for DNA. See Fig. 2g for anti-DNA/DAPI images treated samples (n = 3). g, TBK1/NDP52 DKO cells rescued with GFP–TBK1 wild-type or K38M, or GFP–OPTN(S177D) and were assessed by immunoblotting. h, Cells in g were assessed by immunoblotting. i, Quantification of COXII levels in h displayed as mean ± s.d. from three independent experiments. **P < 0.005 (one-way ANOVA). Scale bars, 10 μm.
Extended Data Figure 4 Parkin-independent recruitment of receptors to mitochondria through PINK1 activity.
a, Isolated mitochondria from wild-type and pentaKO cells with or without FRB-Fis1 and with wild-type or kinase-dead PINK1Δ110–YFP–2×FKBP were immunoblotted. b–e, Representative images of pentaKO cells expressing FRB-Fis1, wild-type or kinase-dead PINK1Δ110–YFP–2×FKBP, and mCh–OPTN or mCh–NDP52 (b), mCh–p62 (c), mCh–OPTN(D474N) (d) or mCh–NDP52(ΔZF) (e). Cells were untreated (b) or treated with rapalog (c–e) and immunostained for TOM20. All images are representative of three independent experiments. See Fig. 3b for quantification. Scale bars, 10 μm.
Extended Data Figure 5 PINK1 directly stimulates mitophagy in the absence of mitochondrial damage.
a, b, Cells were treated with rapalog and analysed by FACS for lysosomal-positive mt-mKeima. Representative data for wild-type HeLa (a) and pentaKO cells (b) without or with Flag/HA–OPTN. c, d, Cell lysates from pentaKO cells expressing FRB-Fis1, PINK1Δ110–YFP–2×FKBP, mt-mKeima and wild-type Flag/HA–OPTN (c) or mutants, (d) Flag/HA–p62, wild-type Flag/HA–NDP52 or NDP52 mutants as indicated were assessed for receptor expression by immunoblotting. e, f, Cells from c and d were treated with rapalog and analysed by FACS for lysosomal-positive mt-mKeima. Representative data of two experiments are presented. g, Cell lysates from pentaKO cells expressing FRB-Fis1, with or without Flag/HA–OPTN and wild-type or kinase-dead PINK1Δ110–YFP–2×FKBP were assessed for OPTN by immunoblotting. h, Flag/HA–OPTN pentaKO cells expressing FRB-Fis1, PINK1Δ110–YFP–2×FKBP, mt-mKeima-transfected and either vector or untagged parkin were analysed by FACS. Representative data of two experiments are presented.
Extended Data Figure 6 PINK1 directly stimulates mitophagy after mitochondrial damage.
a, c, Representative FACS data of mt-mKeima-expressing wild-type or PINK1 knockout HeLa cells (a) or PINK1 knockout cells rescued with PINK1 wild-type and then untreated or treated with oligomycin and antimycin A (c). b, d, Average percentage of mitophagy for two replicates of a and c, respectively. e, Representative images of wild-type HeLa cells expressing mCh–OPTN and treated with oligomycin and antimycin A as indicated and immunostained for TOM20 (n = 3). f, Quantification of mCh–OPTN translocation from cells in e. Data are mean ± s.d. from three independent experiments. ***P < 0.001 (one-way ANOVA). NS, not significant.
Extended Data Figure 7 OPTN and NDP52 preferentially bind phospho-mimetic ubiquitin.
a, HeLa cells expressing mCh–parkin and wild-type HA–ubiquitin (HA–UB) or mutants S65D or S65A were treated with CCCP. HA–ubiquitin was co-immunoprecipitated and the bound fraction was analysed by immunoblotting. Quantifications of the total bound fraction of OPTN, NDP52 and p62 are shown. b, HA–ubiquitin transfected into HeLa cells with mCh–parkin were then treated with CCCP, and HA–ubiquitin was immunoprecipitated. The bound fraction was treated with the deubiquitinase USP2, and washed to remove all unbound protein after deubiquitination. Quantification of the total bound fraction of OPTN, NDP52 and p62 are shown on the right. c, d, Strep-tactin-tagged ubiquitin (strep–UB) was incubated with either wild-type or kinase-dead PINK1 in an in vitro phosphorylation reaction, immunoblotted with an anti-phosphoS65 ubiquitin antibody (c), and then incubated with cytosol collected from untreated, wild-type HeLa cells. The ubiquitin was then pulled down using strep-tactin beads and analysed by immunoblotting (d). e, Quantification of bound OPTN and p62 normalized to total ubiquitin. Data in a, b and e are mean ± s.d. from three independent experiments. *P < 0.05, **P < 0.005, ***P < 0.001 (one-way ANOVA). Dagger symbol denotes nonspecific band.
Extended Data Figure 8 Analysis of LC3 family members and their translocation to damaged mitochondria in autophagy receptor knockout cell lines.
a, Representative images of wild-type, pentaKO and ATG5 knockout HeLa cells expressing mCh–parkin and GFP–LC3B were immunostained for TOM20 (n = 3). b, Cell lysates from mCh–parkin expressing wild-type, pentaKO and ATG5 knockout cells were immunoblotted. c, Representative images of wild-type, NDP52/OPTN DKO and pentaKO cells expressing mCh–parkin and GFP-tagged LC3A, LC3B or LC3C were immunostained for TOM20 (n = 3; see Fig. 4a for quantification). d, e, Representative images of wild-type and NDP52/OPTN/TAX1BP1 triple knockout cells expressing mCh–parkin and GFP–LC3C were immunostained for TOM20 (n = 3) (d) and quantified for GFP–LC3C translocation to mitochondria (e). Data in e are mean ± s.d. from three independent experiments. ***P < 0.001 (one-way ANOVA). Scale bars, 10 μm.
Extended Data Figure 9 GABARAP proteins do not translocate to damaged mitochondria, and early stages of autophagosome biogenesis mediated by WIPI1 and DFCP1 are inhibited in autophagy-receptor-deficient cell lines.
a–c, Representative images of wild-type, NDP52/OPTN DKO and pentaKO cells expressing mCh–parkin and GFP-tagged GABARAP, GABARAPL1 or GABARAPL2 (a), GFP–WIPI1 (b) or GFP–DFCP1 (c) immunostained for TOM20 (n = 3 for each condition, see Fig. 4b and c for quantification of b and c). d, mCh–parkin cell lines as indicated were subjected to either phos-tag SDS–PAGE or standard SDS–PAGE followed by immunoblotting. Arrows indicate the position of phosphorylated beclin species. e, Representative images of untreated wild-type, NDP52/OPTN DKO and pentaKO cell lines expressing mCh–parkin and GFP–ULK1 were immunostained for TOM20 and GFP (n = 3 experiments). Scale bars, 10 μm.
Extended Data Figure 10 OPTN and NDP52 rescue DFCP1 and ULK1 recruitment deficit in pentaKOs.
a, Representative images of pentaKO cells expressing mCh–parkin, GFP–DFCP1 and the indicated Flag/HA-tagged autophagy receptors immunostained for haemagglutinin (n = 2 experiments). Panels on the right display co-localization of Flag/HA-tagged constructs and GFP–DFCP1 by fluorescence intensity line measurement. b, Representative images of pentaKO cells expressing mCh–parkin and GFP–ULK1 that were rescued by Flag/HA–OPTN, Flag/HA–NDP52 and Flag/HA–p62, and immunostained for haemagglutinin and GFP. Arrows indicate HA-tagged receptor puncta (n = 2). Right panels display colocalization of haemagglutinin and GFP by fluorescence intensity line measurement. c, d, Representative images of pentaKO cells stably expressing FRB-Fis1 and transiently expressing PINK1Δ110–YFP–2×FKBP and vector or Myc-tagged receptors, that were untreated (c) or treated with rapalog (d) and imaged live (n = 3 experiments, see Fig. 4h, i for quantification of c, d). Scale bars, 10 μm. e, Old and new models of PINK1/parkin mitophagy. The old model is dominated by parkin ubiquitination of mitochondrial proteins. In this, PINK1 has a small initiator role with the main function being to bring parkin to the mitochondria. The new model depicts parkin-dependent and -independent pathways leading to robust and low-level mitophagy, respectively. On the basis of our data, PINK1 is central to mitophagy both before and after parkin recruitment by phosphorylating ubiquitin to recruit parkin and autophagy receptors to mitochondria, to induce clearance. In the absence of parkin (right), this occurs at a low level owing to the relatively low basal ubiquitin levels on mitochondria. When parkin is present, it serves to amplify the PINK1 generated phospho-ubiquitin signal, allowing for robust and rapid mitophagy induction.
Supplementary information
Supplementary information
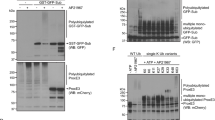
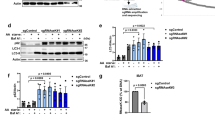
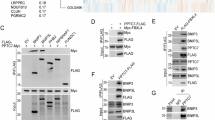
This file contains figures showing the uncropped immunoblots present in the main text and extended data section of the manuscript. (PDF 52015 kb)
Supplementary Tables
This file contains Supplementary Table 1, which shows the genotyping results. (XLSX 44 kb)
Rights and permissions
About this article
Cite this article
Lazarou, M., Sliter, D., Kane, L. et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524, 309–314 (2015). https://doi.org/10.1038/nature14893
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature14893
This article is cited by
-
Selective autophagy in cancer: mechanisms, therapeutic implications, and future perspectives
Molecular Cancer (2024)
-
Panax notoginseng saponins prevent dementia and oxidative stress in brains of SAMP8 mice by enhancing mitophagy
BMC Complementary Medicine and Therapies (2024)
-
Key genes and convergent pathogenic mechanisms in Parkinson disease
Nature Reviews Neuroscience (2024)
-
IKKε and TBK1 prevent RIPK1 dependent and independent inflammation
Nature Communications (2024)
-
Regulation of ULK1 by WTAP/IGF2BP3 axis enhances mitophagy and progression in epithelial ovarian cancer
Cell Death & Disease (2024)