Abstract
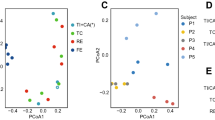
Our knowledge of species and functional composition of the human gut microbiome is rapidly increasing, but it is still based on very few cohorts and little is known about variation across the world. By combining 22 newly sequenced faecal metagenomes of individuals from four countries with previously published data sets, here we identify three robust clusters (referred to as enterotypes hereafter) that are not nation or continent specific. We also confirmed the enterotypes in two published, larger cohorts, indicating that intestinal microbiota variation is generally stratified, not continuous. This indicates further the existence of a limited number of well-balanced host–microbial symbiotic states that might respond differently to diet and drug intake. The enterotypes are mostly driven by species composition, but abundant molecular functions are not necessarily provided by abundant species, highlighting the importance of a functional analysis to understand microbial communities. Although individual host properties such as body mass index, age, or gender cannot explain the observed enterotypes, data-driven marker genes or functional modules can be identified for each of these host properties. For example, twelve genes significantly correlate with age and three functional modules with the body mass index, hinting at a diagnostic potential of microbial markers.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
08 June 2011
An author was omitted. His name has been added to the HTML and PDF and described in the accompanying Corrigendum.
References
Eckburg, P. B. et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005)
Hayashi, H., Sakamoto, M. & Benno, Y. Phylogenetic analysis of the human gut microbiota using 16S rDNA clone libraries and strictly anaerobic culture-based methods. Microbiol. Immunol. 46, 535–548 (2002)
Lay, C. et al. Colonic microbiota signatures across five northern European countries. Appl. Environ. Microbiol. 71, 4153–4155 (2005)
Gill, S. R. et al. Metagenomic analysis of the human distal gut microbiome. Science 312, 1355–1359 (2006)
Turnbaugh, P. J. et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009)
Kurokawa, K. et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14, 169–181 (2007)
Zoetendal, E. G., Rajilic-Stojanovic, M. & de Vos, W. M. High-throughput diversity and functionality analysis of the gastrointestinal tract microbiota. Gut 57, 1605–1615 (2008)
Qin, J. et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65 (2010)
Raes, J. & Bork, P. Molecular eco-systems biology: towards an understanding of community function. Nature Rev. Microbiol. 6, 693–699 (2008)
Nelson, K. E. et al. A catalog of reference genomes from the human microbiome. Science 328, 994–999 (2010)
MetaHIT Consortium . MetaHIT Draft Bacterial Genomes at the Sanger Institute. 〈http://www.sanger.ac.uk/resources/downloads/bacteria/metahit/〉 (9 July 2010)
Muller, J. et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 38, D190–D195 (2010)
Palmer, C., Bik, E. M., Digiulio, D. B., Relman, D. A. & Brown, P. O. Development of the human infant intestinal microbiota. PLoS Biol. 5, e177 (2007)
Tap, J. et al. Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11, 2574–2584 (2009)
Jensen, L. J. et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 37, D412–D416 (2009)
Dethlefsen, L., Huse, S., Sogin, M. L. & Relman, D. A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6, e280 (2008)
Walker, A. Say hello to our little friends. Nature Rev. Microbiol. 5, 572–573 (2007)
Krogfelt, K. A. Bacterial adhesion: genetics, biogenesis, and role in pathogenesis of fimbrial adhesins of Escherichia coli . Rev. Infect. Dis. 13, 721–735 (1991)
Salonen, A. et al. Comparative analysis of fecal DNA extraction methods with phylogenetic microarray: effective recovery of bacterial and archaeal DNA using mechanical cell lysis. J. Microbiol. Methods 81, 127–134 (2010)
Rajilic-Stojanovic, M. et al. Development and application of the human intestinal tract chip, a phylogenetic microarray: analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ. Microbiol. 11, 1736–1751 (2009)
Rousseeuw, P. J. Silhouettes: a graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987)
Vanhoutte, T., Huys, G., Brandt, E., d & Swings, J. Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol. Ecol. 48, 437–446 (2004)
Tannock, G. W. et al. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 66, 2578–2588 (2000)
Seksik, P. et al. Alterations of the dominant faecal bacterial groups in patients with Crohn’s disease of the colon. Gut 52, 237–242 (2003)
Costello, E. K. et al. Bacterial community variation in human body habitats across space and time. Science 326, 1694–1697 (2009)
Martens, E. C., Koropatkin, N. M., Smith, T. J. & Gordon, J. I. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J. Biol. Chem. 284, 24673–24677 (2009)
Wright, D. P., Rosendale, D. I. & Roberton, A. M. Prevotella enzymes involved in mucin oligosaccharide degradation and evidence for a small operon of genes expressed during growth on mucin. FEMS Microbiol. Lett. 190, 73–79 (2000)
Derrien, M., Vaughan, E. E., Plugge, C. M. & de Vos, W. M. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int. J. Syst. Evol. Microbiol. 54, 1469–1476 (2004)
Ley, R. E., Turnbaugh, P. J., Klein, S. & Gordon, J. I. Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023 (2006)
Schwiertz, A. et al. Microbiota and SCFA in lean and overweight healthy subjects. Obesity 18, 190–195 (2009)
Woodmansey, E. J. Intestinal bacteria and ageing. J. Appl. Microbiol. 102, 1178–1186 (2007)
Kovacikova, G. & Skorupski, K. The alternative sigma factor σE plays an important role in intestinal survival and virulence in Vibrio cholerae . Infect. Immun. 70, 5355–5362 (2002)
Fujihashi, K. & Kiyono, H. Mucosal immunosenescence: new developments and vaccines to control infectious diseases. Trends Immunol. 30, 334–343 (2009)
Turnbaugh, P. J. et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006)
Raes, J., Korbel, J. O., Lercher, M. J., von Mering, C. & Bork, P. Prediction of effective genome size in metagenomic samples. Genome Biol. 8, R10 (2007)
Gibson, G. R. et al. Alternative pathways for hydrogen disposal during fermentation in the human colon. Gut 31, 679–683 (1990)
Godon, J. J., Zumstein, E., Dabert, P., Habouzit, F. & Moletta, R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 63, 2802–2813 (1997)
Arumugam, M., Harrington, E. D., Foerstner, K. U., Raes, J. & Bork, P. Smash Community: a metagenomic annotation and analysis tool. Bioinformatics 26, 2977–2978 (2010)
Wang, Q., Garrity, G. M., Tiedje, J. M. & Cole, J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007)
Gianoulis, T. A. et al. Quantifying environmental adaptation of metabolic pathways in metagenomics. Proc. Natl Acad. Sci. USA 106, 1374–1379 (2009)
Acknowledgements
The authors are grateful to C. Creevey, G. Falony and members of the Bork group at EMBL for discussions and assistance. We thank the EMBL IT core facility and Y. Yuan for managing the high-performance computing resources. The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007-2013): MetaHIT, grant agreement HEALTH-F4-2007-201052, EMBL, the Lundbeck Foundation Centre for Applied Medical Genomics in Personalized Disease Prediction, Prevention and Care (LuCAMP), Novo Nordisk Foundation and the International Science and Technology Cooperation Project in China (0806). Obese/non-obese volunteers for the MicroObes study were recruited from the SU.VI.MAX cohort study coordinated by P. Galan and S. Hercberg, and metagenome sequencing was funded by Agence Nationale de la Recherche (ANR); volunteers for MicroAge study were recruited from the CROWNALIFE cohort study coordinated by S. Silvi and A. Cresci, and metagenome sequencing was funded by GenoScope. Ciberehd is funded by the Instituto de Salud Carlos III (Spain). J.R. is supported by the Institute for the encouragement of Scientific Research and Innovation of Brussels (ISRIB) and the Odysseus programme of the Fund for Scientific Research Flanders (FWO). We are thankful to the Human Microbiome Project for generating the reference genomes from human gut microbes and the International Human Microbiome Consortium for discussions and exchange of data.
Author information
Authors and Affiliations
Consortia
Contributions
All authors are members of the Metagenomics of the Human Intestinal Tract (MetaHIT) Consortium. Jun W., F.G., O.P., W.M.d.V., S.B., J.D., Jean W., S.D.E. and P.B. managed the project. N.B., F.C., T.H., C.M. and T. N. performed clinical analyses. M.L. and F.L. performed DNA extraction. E.P., D.L.P., T.B., J.P. and E.U. performed DNA sequencing. M.A., J.R., S.D.E. and P.B. designed the analyses. M.A., J.R., T.Y., D.R.M., G.R.F., J.T., J.-M.B., M.B., L.F., L.G., M.K., H.B.N., N.P., J.Q., T.S.-P., S.T., D.T., E.G.Z., S.D.E. and P.B. performed the analyses. M.A., J.R., P.B. and S.D.E. wrote the manuscript. M.H., T.H., K.K. and the MetaHIT Consortium members contributed to the design and execution of the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Raw Sanger read data from the European faecal metagenomes have been deposited in the NCBI Trace Archive with the following project identifiers: MH6 (33049), MH13 (33053), MH12 (33055), MH30 (33057), CD1 (33059), CD2 (33061), UC4 (33113), UC6 (33063), NO1 (33305), NO3 (33307), NO4 (33309), NO8 (33311), OB2 (33313), OB1 (38231), OB6 (38233), OB8 (45929), A (63073), B (63075), C (63077), D (63079), E (63081), G (63083). Contigs, genes and annotations are available to download from http://www.bork.embl.de/Docu/Arumugam_et_al_2011/.
Supplementary information
Supplementary Information
The file contains Supplementary Methods, Supplementary Notes and Supplementary References. A minor error in Supplementary Information section 2.2 was corrected on 02 June 2011. (PDF 769 kb)
Supplementary Figures
This file contains Supplementary Figures 1-27 with legends. (PDF 3115 kb)
Supplementary Tables
The file contains Supplementary Tables 1 - 2 and 4 - 24 (see separate file for Supplementary Table 3). (PDF 520 kb)
Supplementary Table 3
The file contains Supplementary Table 3. (PDF 1175 kb)
Rights and permissions
About this article
Cite this article
Arumugam, M., Raes, J., Pelletier, E. et al. Enterotypes of the human gut microbiome. Nature 473, 174–180 (2011). https://doi.org/10.1038/nature09944
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature09944
This article is cited by
-
The role of gut microbiota in human metabolism and inflammatory diseases: a focus on elderly individuals
Annals of Microbiology (2024)
-
Faecalibacterium prausnitzii as a potential Antiatherosclerotic microbe
Cell Communication and Signaling (2024)
-
Identification of enterotype and its predictive value for patients with colorectal cancer
Gut Pathogens (2024)
-
Unlocking the secrets: exploring the influence of the aryl hydrocarbon receptor and microbiome on cancer development
Cellular & Molecular Biology Letters (2024)
-
Exploring the presence of oral bacteria in non-oral sites of patients with cardiovascular diseases using whole metagenomic data
Scientific Reports (2024)