Abstract
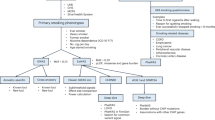
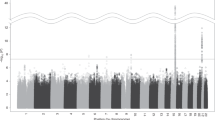
The common nonsynonymous variant rs16969968 in the α5 nicotinic receptor subunit gene (CHRNA5) is the strongest genetic risk factor for nicotine dependence in European Americans and contributes to risk in African Americans. To comprehensively examine whether other CHRNA5 coding variation influences nicotine dependence risk, we performed targeted sequencing on 1582 nicotine-dependent cases (Fagerström Test for Nicotine Dependence score⩾4) and 1238 non-dependent controls, with independent replication of common and low frequency variants using 12 studies with exome chip data. Nicotine dependence was examined using logistic regression with individual common variants (minor allele frequency (MAF)⩾0.05), aggregate low frequency variants (0.05>MAF⩾0.005) and aggregate rare variants (MAF<0.005). Meta-analysis of primary results was performed with replication studies containing 12 174 heavy and 11 290 light smokers. Next-generation sequencing with 180 × coverage identified 24 nonsynonymous variants and 2 frameshift deletions in CHRNA5, including 9 novel variants in the 2820 subjects. Meta-analysis confirmed the risk effect of the only common variant (rs16969968, European ancestry: odds ratio (OR)=1.3, P=3.5 × 10−11; African ancestry: OR=1.3, P=0.01) and demonstrated that three low frequency variants contributed an independent risk (aggregate term, European ancestry: OR=1.3, P=0.005; African ancestry: OR=1.4, P=0.0006). The remaining 22 rare coding variants were associated with increased risk of nicotine dependence in the European American primary sample (OR=12.9, P=0.01) and in the same risk direction in African Americans (OR=1.5, P=0.37). Our results indicate that common, low frequency and rare CHRNA5 coding variants are independently associated with nicotine dependence risk. These newly identified variants likely influence the risk for smoking-related diseases such as lung cancer.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Dani JA, De Biasi M . Cellular mechanisms of nicotine addiction. Pharmacol Biochem Behav 2001; 70: 439–446.
Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nat Genet 2008; 40: 616–622.
Berrettini W, Yuan X, Tozzi F, Song K, Francks C, Chilcoat H et al. Alpha-5/alpha-3 nicotinic receptor subunit alleles increase risk for heavy smoking. Mol Psychiatry 2008; 13: 368–373.
Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X et al. Variants in nicotinic receptors and risk for nicotine dependence. Am J Psychiatry 2008; 165: 1163–1171.
Caporaso N, Gu F, Chatterjee N, Sheng-Chih J, Yu K, Yeager M et al. Genome-wide and candidate gene association study of cigarette smoking behaviors. PLoS One 2009; 4: e4653.
Chen LS, Baker TB, Piper ME, Breslau N, Cannon DS, Doheny KF et al. Interplay of genetic risk factors (CHRNA5-CHRNA3-CHRNB4) and cessation treatments in smoking cessation success. Am J Psychiatry 2012; 169: 735–742.
Hung RJ, McKay JD, Gaborieau V, Boffetta P, Hashibe M, Zaridze D et al. A susceptibility locus for lung cancer maps to nicotinic acetylcholine receptor subunit genes on 15q25. Nature 2008; 452: 633–637.
Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet 2009; 5: e1000421.
Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Hum Mol Genet 2007; 16: 36–49.
Thorgeirsson TE, Geller F, Sulem P, Rafnar T, Wiste A, Magnusson KP et al. A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 2008; 452: 638–642.
Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet 2010; 42: 436–440.
Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet 2010; 42: 448–453.
Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet 2010; 42: 441–447.
Chen LS, Saccone NL, Culverhouse RC, Bracci PM, Chen CH, Dueker N et al. Smoking and genetic risk variation across populations of European, Asian, and African American ancestry—a meta-analysis of chromosome 15q25. Genet Epidemiol 2012; 36: 340–351.
Saccone NL, Wang JC, Breslau N, Johnson EO, Hatsukami D, Saccone SF et al. The CHRNA5-CHRNA3-CHRNB4 nicotinic receptor subunit gene cluster affects risk for nicotine dependence in African-Americans and in European-Americans. Cancer Res 2009; 69: 6848–6856.
Kuryatov A, Berrettini W, Lindstrom J . Acetylcholine receptor (AChR) alpha5 subunit variant associated with risk for nicotine dependence and lung cancer reduces (alpha4beta2)alpha5 AChR function. Mol Pharmacol 2011; 79: 119–125.
Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet 2007; 16: 24–35.
Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO . The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. Br J Addict 1991; 86: 1119–1127.
Wang K, Li M, Hakonarson H . ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010; 38: e164.
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012; 491: 56–65.
Gonzalez-Perez A, Lopez-Bigas N . Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet 2011; 88: 440–449.
Haller G, Druley T, Vallania FL, Mitra RD, Li P, Akk G et al. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Hum Mol Genet 2012; 21: 647–655.
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D . Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 2006; 38: 904–909.
Li B, Leal SM . Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet 2008; 83: 311–321.
Nagelkerke N . A note on a general definition of the coefficient of determination. Biometrika 1991; 78: 691–692.
Stevens VL, Bierut LJ, Talbot JT, Wang JC, Sun J, Hinrichs AL et al. Nicotinic receptor gene variants influence susceptibility to heavy smoking. Cancer Epidemiol Biomarkers Prev 2008; 17: 3517–3525.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007; 81: 559–575.
Doyle GA, Chou AD, Saung WT, Lai AT, Lohoff FW, Berrettini WH . Identification of CHRNA5 rare variants in African-American heavy smokers. Psychiatr Genet 2014; 24: 102–109.
Vrieze SI, Feng S, Miller MB, Hicks BM, Pankratz N, Abecasis GR et al. Rare nonsynonymous exonic variants in addiction and behavioral disinhibition. Biol Psychiatry 2014; 75: 783–789.
Wessel J, McDonald SM, Hinds DA, Stokowski RP, Javitz HS, Kennemer M et al. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerstrom test for nicotine dependence. Neuropsychopharmacology 2010; 35: 2392–2402.
Maher B . Personal genomes: The case of the missing heritability. Nature 2008; 456: 18–21.
Keskitalo K, Broms U, Heliovaara M, Ripatti S, Surakka I, Perola M et al. Association of serum cotinine level with a cluster of three nicotinic acetylcholine receptor genes (CHRNA3/CHRNA5/CHRNB4) on chromosome 15. Hum Mol Genet 2009; 18: 4007–4012.
Bloom AJ, Hartz SM, Baker TB, Chen LS, Piper ME, Fox L et al. Beyond cigarettes per day. A genome-wide association study of the biomarker carbon monoxide. Ann Am Thorac Soc 2014; 11: 1003–1010.
Munafo MR, Timofeeva MN, Morris RW, Prieto-Merino D, Sattar N, Brennan P et al. Association between genetic variants on chromosome 15q25 locus and objective measures of tobacco exposure. J Natl Cancer Inst 2012; 104: 740–748.
Wang JC, Cruchaga C, Saccone NL, Bertelsen S, Liu P, Budde JP et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Hum Mol Genet 2009; 18: 3125–3135.
Wang JC, Grucza R, Cruchaga C, Hinrichs AL, Bertelsen S, Budde JP et al. Genetic variation in the CHRNA5 gene affects mRNA levels and is associated with risk for alcohol dependence. Mol Psychiatry 2009; 14: 501–510.
Wang JC, Spiegel N, Bertelsen S, Le N, McKenna N, Budde JP et al. Cis-regulatory variants affect CHRNA5 mRNA expression in populations of African and European ancestry. PLoS One 2013; 8: e80204.
Saccone NL, Schwantes-An TH, Wang JC, Grucza RA, Breslau N, Hatsukami D et al. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes Brain Behav 2010; 9: 741–750.
Acknowledgements
This work was supported by the National Institutes of Health: grant numbers T32GM07200, UL1TR000448, TL1TR000449 and F30AA023685 to EO; grant numbers K08 DA030398 and R01 DA038076 to LC; and grant number U19CA148172 to LJB. Grant number R01 HL118305 from the National Institutes of Health supported the replication analyses. Grants and contracts from the National Institutes of Health supported the following studies and groups: COGEND (P01CA89392), AAND (R01DA025888), CIDR (HHSN268201100011I), ARIC (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C, HHSN268201100012C, R01HL087641, R01HL59367, R01HL086694, U01HG004402, HHSN268200625226C, UL1RR025005, 5RC2HL102419), CHS (HHSN268201200036C, HHSN268200800007C, N01HC55222, N01HC85079, N01HC85080, N01HC85081, N01HC85082, N01HC85083, N01HC85086, U01HL080295, R01HL087652, R01HL105756, R01HL103612, R01HL120393, R01HL085251, R01HL068986, R01AG023629, UL1TR000124, DK063491), COGA (U10AA008401), FamHS (R01HL118305, R01DK089256), GENOA (HL054464, HL054457, HL054481, HL071917, NS041558, HL87660, HL119443, HL118305), HyperGEN (HL54471, HL54472, HL54473, HL54495, HL54496, HL54497, HL54509, HL54515, R01HL55673, R01HL055673, R01HL118305, U01HL54473, R01HL055673, R01HL118305), JHS (HSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN268201300050C, HL103010, HL118305), MESA (N01HC95159, N01HC95160, N01HC95161, N01HC95162, N01HC95163, N01HC95164, N01HC95165, N01HC95166, N01HC95167, N01HC95168, N01HC95169, UL1TR000040, UL1RR025005, R01HL071051, R01HL071205, R01HL071250, R01HL071251, R01HL071252, R01HL071258, R01HL071259, UL1RR025005, N02HL64278, UL1TR000124, DK063491), WGHS (HL043851, HL080467, CA047988), WHI (HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C, HHSN271201100004C, R21HL123677, R01HL118305). Erasmus Rucphen Family Study was supported by the following grants: European Commission FP6 STRP grant number 018947 (LSHG-CT-2006-01947); European Community's Seventh Framework Program (FP7/2007–2013, HEALTH-F4-2007-201413); Netherlands Organization for Scientific Research and the Russian Foundation for Basic Research (NWO-RFBR 047.017.043); ZonMw grant (project 91111025). Rotterdam Study was supported by Netherlands Organisation of Scientific Research NWO Investments (nr. 175.010.2005.011, 911-03-012). This study was also funded by the Research Institute for Diseases in the Elderly (014-93-015; RIDE2), the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) project nr. 050-060-810 and Netherlands Consortium for Healthy Ageing (NCHA). Please see Supplementary Materials for acknowledgements listed by study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
LJB, AG and J-CW as well as the spouse of NLS are listed as inventors on Issued U.S. Patent 8,080,371, ‘Markers for Addiction’ covering the use of certain SNPs in determining the diagnosis, prognosis and treatment of addiction. JAS has received support from Pfizer, Inc. NA is supported by the Hersenstichting Nederland (project number F2013(1)-28). OHF works in ErasmusAGE, a center for aging research across the life course funded by Nestlé Nutrition (Nestec Ltd.), Metagenics Inc. and AXA. Nestlé Nutrition (Nestec Ltd.), Metagenics Inc. and AXA had no role in design and conduct of the study; collection, management, analysis and interpretation of the data; and preparation, review or approval of the manuscript. BMP serves on the DSMB of a clinical trial funded by the device manufacturer (Zoll LifeCor) and on the Steering Committee of the Yale Open Data Access Project funded by Johnson & Johnson.
Additional information
Supplementary Information accompanies the paper on the Molecular Psychiatry website
Supplementary information
PowerPoint slides
Rights and permissions
About this article
Cite this article
Olfson, E., Saccone, N., Johnson, E. et al. Rare, low frequency and common coding variants in CHRNA5 and their contribution to nicotine dependence in European and African Americans. Mol Psychiatry 21, 601–607 (2016). https://doi.org/10.1038/mp.2015.105
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/mp.2015.105