Abstract
Reproducibility of computational research is often challenging despite established guidelines and best practices. Translating these guidelines into practical applications remains difficult. Here, we present ENCORE, an approach to enhance transparency and reproducibility by guiding researchers in how to structure and document a computational project. ENCORE builds on previous efforts in computational reproducibility and integrates all project components into a standardized file system structure. It utilizes pre-defined files as documentation templates, leverages GitHub for software versioning, and includes an HTML-based navigator. ENCORE is designed to be agnostic to the type of computational project, data, programming language, and ICT infrastructure, and does not rely on specific software tools. We also share our group’s experience using ENCORE, highlighting that the most significant challenge to the routine adoption of approaches like ours is the lack of incentives to motivate researchers to dedicate sufficient time and effort to ensure reproducibility.
Similar content being viewed by others
Introduction
Reproducibility of research outcomes has become increasingly important within all research fields, including the (bio)medical domain1,2. The scientific community is increasingly concerned about the so-called ‘reproducibility crisis’, which includes but is not limited to the failure to reproduce results of published studies and lack of transparency and completeness3. Research transparency importantly contributes to reproducibility and refers to the degree to which methodologies, data, and results are accessible and understandable to others and, consequently, contributes to the overall credibility and trustworthiness of a study4. Increased data size, increased complexity of experimental and computational methods, and multi-disciplinarity, make reproducibility increasingly challenging. Scientist-driven efforts complemented with pressure from research institutes, funders, and journals have resulted in various initiatives and approaches, covering the different stages of the research lifecycle, to increase reproducibility of scientific findings5 (Fig. 1). Typically, this cycle starts (stage 1) with a careful preparation of the study, which includes planning in terms of research objectives, funding, data management, ethics, experimental design, experimental and computational approaches, publishing, and study archiving. Stage 2 concerns the collection and processing of (patient) samples, and the generation of data. Once data has been collected, it is analyzed using statistical or other computational methods (stage 3). This stage may include, for example, quality control of experimental data, evaluation of data analysis, mathematical modeling, and simulations depending on the research project. Alternatively, one may solely or partially rely on existing data from (public) repositories or conduct studies that are based on computer simulations6,7. Stage 4 (publication and archiving) is increasingly driven by initiatives aiming for open-access publications and data8,9 and open-source software10,11. Research findings are published in open-access preprint repositories (e.g., arXiv, bioRxiv, medRxiv) and/or peer-reviewed (open-access) journals, while additional output (e.g., data, code) can be archived in, for example, the major omics repositories12, figshare13 and Zenodo14.
Most (biomedical) studies go through the stages of (1) study preparation, (2) data collection (involving sample collection and processing, and data generation), (3) computational approaches for data analyzes, modeling and/or simulations, and (4) (open-access/open-source) publication and archiving. Published research findings drive new studies. ENCORE focuses on computational reproducibility (stage 3).
Despite ongoing initiatives to improve computational reproducibility, results from computational analyzes, including from our group, often remain difficult to reproduce even if data and code are available15,16,17,18. Irreproducibility of computational research has well-known causes, including undocumented manual processing steps, unavailability of software or data, reliance on public repositories, changes in web-services and software libraries, incomplete software documentation, and unspecified parameters. Guidelines to improve computational reproducibility have been suggested by several groups (e.g.19,20,21) but their translation and application in research practice seems to be a bottleneck given that computational studies do not always (fully) adhere to these guidelines. Reproducibility may also be compromised if software is not transparent and, consequently, is not easy to run, understand, test, or modify. This problem typically arises in software developed by (biomedical) researchers without formal training in software engineering22,23. Such software might even contain undetected (conceptual) errors although this does not necessarily result in irreproducible results. Nevertheless, training researchers with best practices in software engineering is expected to improve reproducibility. Moreover, making software available as open-source24, allows peers to inspect, use, verify, correct, and/or extend the software. This prompts for the use of code versioning systems like Git and GitHub25 or alternatives like GitLab and BitBucket, which also facilitate code hosting and sharing, and allow collaborative software development thereby supporting transparency and reproducibility of computational research26,27. Note however that code versioning systems are not necessarily persistent but that solutions are in place for example by authorizing Zenodo to archive a specific release of a GitHub repository and issue a DOI for this archive.
Over the last decade, FAIR (Findable, Accessible, Interoperable, Reusable) data management has been introduced to ensure the availability and reusability of experimental data28 while newly developed software is increasingly made available on GitHub and other dedicated software repositories11,29. However, in our view, the separation of data and software in different repositories is another potential cause for irreproducibility since manual steps may be required to link data and software. Since such steps are generally not described in published papers, it is left to the researcher to reconstruct the precise computational workflow. Moreover, by storing code and data in different repositories, a direct connection with (published) results is also lost. The lack of detailed project documentation further impedes transparency and reproducibility. Therefore, we believe that computational reproducibility is improved if data, code, and results are more tightly integrated and well-documented. Such a self-contained project compendium could also more easily be shared with peers and reviewers.
In this paper, we focus on computational reproducibility, that is, the reanalysis of the same data using the same computational methods. We propose ENCORE (ENhancing COmputational REproducibility)30 as a practical approach to improve transparency and reproducibility. The approach is based on eight main requirements that we define below (see Methods), and four practical principles (Supplementary Method 1). The development of ENCORE started in 2018 and is based on existing ideas and initiatives in particular from Spreckelsen and co-workers31 for using a standardized way to organize research projects, but also other initiatives5,19,21,27,32,33,34,35,36,37,38,39, discussions within our research group, and improvements made over a five-year period. It aims to integrate all project components in a single project compendium. ENCORE substantially improves transparency and reproducibility and demonstrates how the eight requirements and four principles are met in practice. ENCORE also guides researchers to structure and document the computational part of a research project. It harmonizes the approach towards reproducibility if adopted by a whole research group, which brings additional advantages. ENCORE does not specifically address stages 1, 2 and 4 of the research lifecycle (Fig. 1). However, (parts of) the documentation resulting from these stages will generally be included in the project compendium not only for completeness but also to correctly design and perform the computational analyzes40. This includes but is not limited to information about hypotheses to be tested, experimental design, descriptions of (patient) samples and the physical experiment, and the (FAIR) data. Conversely, the ENCORE documentation of computational protocols provides information for annotating the (pre)processed data to meet the FAIR guiding principles. The ENCORE compendium can serve as supplementary information for research papers. It is, however, important to recognize that ENCORE will generally contain much more information than published in a typical research paper. Here, we describe the design of ENCORE, our experiences, and results from internal evaluations. ENCORE improved reproducibility and transparency of our computational projects but, at the same time, made clear that achieving full reproducibility will require further steps. Perhaps the most significant challenge to overcome for routine usage of initiatives such as ENCORE is the lack of incentives to motivate researchers to spend sufficient time and effort on reproducibility. We expect ENCORE to contribute to the further development of approaches to increase transparency and reproducibility of computational research.
Results
Initial discussions and review of publications to improve reproducibility of research projects in our group started in 2018 (Fig. 2), and consistently involved all group members (i.e., staff, postdocs, PhD students, and internship students). This led to the aims specified in the introduction (transparency, reproducibility, harmonization) and to a first version of ENCORE in October 2020. At the same time, we initiated a GitHub organization account41 to host all ENCORE repositories from our group. The use of ENCORE was made mandatory for all new research projects and for everyone in our group. This initial step led to the harmonization of project organization within our group. Currently, we have over 20 ENCORE research projects. In addition, we have over 50 ENCORE projects for data analysis services provided to other groups.
ENCORE evolved through different versions incorporating changes based on practical experiences, evaluations, and group discussions. This led to gradual improvements in the ENCORE approach and documentation, broader use for research projects, and Git/GitHub proficiency within our research group. In turn, this led to better and more transparent organization of projects and increased reproducibility. Further improvements and extensions are expected in the future.
Evolution and evaluation
ENCORE is based on a standardized file system structure (sFSS) including pre-defined files to guide project documentation. The sFSS serves as a self-contained project compendium. Based on experience with ENCORE for a still increasing number of projects, internal evaluations, and frequent discussions about the structure, pre-defined files, and usage guidelines, we gradually improved the initial setup. Overall, over the past four years, this led to a simplification of the sFSS structure, a reduced number of pre-defined files, and better ENCORE instructions and documentation (see also Supplementary Fig. 2 in Supplementary Methods 1).
A first evaluation was performed one month after introducing ENCORE 1.0 to exchange user experiences, which led to ENCORE 2.0. About three months later, in January 2021, we organized a second evaluation, during which each group member presented an ENCORE project. This evaluation made clear that most research projects adopted the sFSS structure, but that documentation (e.g., README files, lab journal) was occasionally incomplete or missing since keeping documentation up to date was perceived as overhead and time-consuming but also because it was often unclear what level of detail needed to be provided since no clear objectives were set. Some projects did not fully adhere to the sFSS naming and structure conventions, which was partially due to unclear or implicit ENCORE guidelines but also because some group members were still inclined to stick to old habits. Based on this evaluation we developed ENCORE 3.0, which comprised further simplifications of the directory structure (fewer directory levels), renaming of pre-defined files to improve consistency, and improved documentation (i.e., usage rules). In September 2022, we evaluated ENCORE 3.0 to test if ENCORE had indeed improved the reproducibility of our projects. We selected nine ENCORE projects, which were assigned to group members not involved in that project. Subsequently, each project was evaluated for the correct use of the sFSS, the pre-defined files and the level of documentation. In addition, each group member was asked to reproduce a specific subset of project results. The most important outcome of this evaluation was that only for about half of the selected projects the results could be reproduced. Various reasons prevented (full) reproducibility such as the use of different library versions used by the software, use of absolute directory paths that were not valid after transferring the sFSS to a different location, differences between operating systems, lack of instructions on which software to install and how to run the software, compilation problems, software errors, or difficulties in handling large datasets. In addition, some of the projects were difficult to understand due to lack of documentation (transparency) about goals and applied methodology. Some of these problems can easily be fixed such as changing absolute to relative directory paths. Other problems such as dealing with different library versions require more effort to solve (see Discussion). Several other outcomes resulted from this evaluation. First, not being acquainted with a specific project, it sometimes was difficult to find the (core) information that is needed to reproduce and understand a project. This triggered the development of the sFSS Navigator42, which was implemented as an R application in ENCORE 3.1 and later re-implemented in Python and extended in ENCORE 3.5. Second, despite more than two years of discussion and joint decision making about the structure of ENCORE, there was still debate about (how to use) the sFSS structure and the pre-defined files. The sFSS structure and files had been agreed upon by consensus, but still was a compromise of different structures previously used by individual group members. As a result, some parts of it seemed illogical to some members, who ideally still preferred a different setup. In addition, although the ENCORE user guidelines and their rationales had been discussed, agreed upon, and written down, it turned out to be difficult to memorize and apply all these rules. In our view, this is also one of the main reasons that existing published guidelines are not consistently applied. At the same time, the user documentation was scattered over many files and had lost consistency over the evolution of ENCORE. This prompted the changes incorporated in ENCORE 3.5 and 4.0. Specifically, these include the completion of the Step-by-Step ENCORE Guide (Supplementary Method 1) and the merger of documentation and templates directly in the README files (e.g., Supplementary Method 3). Third, the provided level of detail for the documentation of most projects was inadequate, partially caused by unclear guidelines. For example, it was not always clear how to run the software because our guidelines didn’t make explicit how this should be documented and at what level of detail. One specific issue was that the lab journal often did not contain an adequate summary of (supervisory) meetings and email exchanges. It was therefore agreed that it is the responsibility of the project team to ensure that all relevant documentation is incorporated in the sFSS. Finally, it became clear that project organization and documentation often had lower priority than doing the actual research. For these reasons, most projects did not fully adhere to the ENCORE guidelines. Nevertheless, we decided to keep the approach mandatory and to only make minor changes to the current template to prevent delay in its further development.
Using remote computer systems with ENCORE
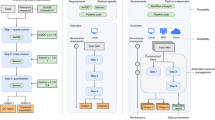
Most of our ENCORE projects reside in the cloud (e.g., the Dutch SURFdrive, Dropbox) and are synchronized with the local computers (e.g., laptop, desktop PC) of the project participants. However, for part of our projects we use dedicated high performance computing infrastructure such as computer clusters to perform computations. The sFSS is compatible with such a scenario. One may simply transfer (part of) the sFSS to the remote system and, subsequently, perform the calculations there. Results can then be transferred back to the cloud and local computer (Fig. 3).
A specific ENCORE project that contains code for data pre-processing and a simulation. An ENCORE project may reside on a local computer (e.g., laptop, desktop PC), on a remote (cloud) computer server such as provided by Amazon Web Services or Dropbox for easy sharing between project participants. All or part of a project compendium can also be temporarily transferred to a HPC facility to perform CPU intensive calculations and transferred back with results once calculations are finished. In this example, only the simulation branch is transferred to a HPC facility. To transfer files to a remote computer system, one may use common data transfer tools, such as curl or rclone that support many data transfer protocols such as sFTP and SCP.
Discussion
We presented ENCORE as a practical implementation, based on eight main requirements (Methods) and four practical principles (Supplementary Method 1, Section 2), guiding researchers on how to structure and document computational projects according to published guidelines5,19,31,32 in order to improve transparency and reproducibility, and to improve harmonization within and across research groups. ENCORE does not consider replicability (sometimes referred to as repeatability), which is about strengthening scientific evidence from replication studies by other research groups using independent data, and experimental and computational methods43,44,45,46. An important aspect of ENCORE is the integration of all project information (concepts, data, code, results, documentation) in a single directory structure that can easily be shared and archived. Although we didn’t have a pre-ENCORE baseline measurement for reproducibility in our group, ENCORE harmonized the way we work in a broad range of projects and provided a big step forward in terms of the organization, transparency, and reproducibility of our projects. Integration and documentation to achieve transparency have also been referred to as the third dimension of open science47 and are in our view key to reproducibility of computational research.
The most important lessons learned from ENCORE are summarized by the following five points:
-
A significant barrier to enhancing transparency and reproducibility is the lack of incentives for researchers to invest sufficient time and effort in these areas.
-
Successful implementation requires incremental steps to minimize disruption to the current ways of working of individual researchers, along with regular discussions and evaluations to monitor and maintain progress.
-
Harmonizing the approach within a research group facilitates the joint development of best practices for reproducibility and makes the inspection and use of projects from colleagues much easier.
-
The next version of ENCORE should explicitly incorporate best practices for software engineering and methods to preserve the computing environment.
-
Further development of ENCORE should involve research groups from diverse domains for further evaluation, improvement, and extension.
Although ENCORE has been developed and tested from the perspective of bioinformatics and computational modeling in the cellular and molecular biology field, it can be applied in other scientific disciplines to virtually any type of computational project: it is agnostic to the computational infrastructure, and it can be used with any programming language or software tool the researcher is using. We emphasize, however, that ENCORE does not neglect existing tools that contribute to the further improvement of reproducibility. ENCORE users are encouraged to utilize complementary tools that enhance reproducibility, including those for (i) preservation of the compute environment, (ii) software development, (iii) workflow management, (iv) (software) documentation, and (v) project management. ENCORE does not impose these tools on researchers; instead, it allows them to choose the most suitable options for their specific needs (Supplementary Method 4). However, for many researchers, changing their individual and favored project organization to the ENCORE sFSS may provide a barrier. Currently, we are taking initial steps to evaluate and further develop ENCORE in collaboration with other research groups. For this, the ENCORE documentation is essential for introducing new ENCORE users to the underlying philosophy and approach and to provide background on the structure and desired level of project documentation. Over the past few years, we have experienced that it is crucial to use ENCORE from the start of a project and to have sufficient self-discipline to keep everything up to date. For project documentation, we follow the guiding principle that it should be at a level of detail that one’s peer or supervisor should be able to understand all aspects of the project (concepts, data, code, results). Early versions of (prototype) code, trial results from data analyzes, etc. that are not expected to be kept in the final compendium can be excluded from documentation to minimize overhead. Nevertheless, in practice, the level of documentation detail often leads to discussion and over the years has led to changes in the instructions and template in the README files. In the future, emerging AI-based tools might assist to automatically generate project documentation from, for example, rough notes, or audio/video recordings of project meetings.
Different areas of reproducibility have been distinguished by Stodden40. (i) Empirical reproducibility refers to physical experiments and the (reporting) standards associated with these. (ii) Computational reproducibility is concerned with the reproduction of results using the same data, computational methodology, and software versions. (iii) Statistical reproducibility focuses on the correct use of experimental design (including sample size calculations) and statistical analyzes. (iv) Ethical reproducibility refers to reporting ethics methods in biomedical research48. ENCORE focuses on computational reproducibility and statistical reproducibility, at the same time considering that documentation about the physical experiments can be important for the computational analyzes. Ethical reproducibility may come into play for specific (artificial intelligence) applications e.g.49, but currently is not explicitly considered by ENCORE.
ENCORE promotes a pre-defined directory structure, integration of data, methods, and results, and detailed project documentation. This supports the continuity of research lines which are sometimes compromised by the continuous flux of scientific personnel in academic groups. ENCORE contributes to transparency and, as such, helps to detect software errors and conceptual methodological flaws by (external) researchers, supervisor(s), and reviewers. Transparency allows project supervisors to provide timely and more constructive feedback. Co-authors of a manuscript can more easily inspect details of the project before manuscript submission to, for example, comply with the ICMJE authorship rules50. A recent editorial in Nature Human Behavior51 proposed that software be part of the peer review process. ENCORE would facilitate such efforts since, in principle, it provides user documentation to install and use the software and datasets. Generally, it would be very time-consuming and difficult to check the code itself, but reviewers could at least check if the results presented in a paper can be reproduced by executing the software.
Increasing reproducibility, what is the problem? The main hurdle to increasing the reproducibility of computational projects is neither the lack of guidelines nor substantial technical barriers. However, despite all discussions and initiatives concerning reproducibility, and our personal observation that many researchers agree on the importance of reproducibility, there still is a long way to go. Regularly, during the development of ENCORE, group members brought forward various arguments for not following (ENCORE) guidelines. For example, one argument being that it consumes time while we are virtually never asked by peers, reviewers, or funding agencies whether our research is reproducible. In fact, there is often no penalty for being non-reproducible and, moreover, there also is no clear reward for working reproducibly. Markowetz gives several examples of benefits of working in a reproducible manner35, but he also encountered resistance from researchers when advocating reproducible research such as “I’d rather do real science than tidy up my data”. Indeed, an often-heard argument concerns the amount of overhead that comes along with (ENCORE) reproducibility approaches. However, for a typical research project, the time spent on following the ENCORE approach (e.g., documentation, structuring) is in our view negligible compared to the time spent on the actual research. There are clear advantages of working reproducibly, which has also been argued by other groups (e.g.4,19,52. For example, reproducibility is important for trust in science, it helps writing a publication, it will save time in the long run, and it supports the sustainability of the research. Although all true, for some researchers these arguments do not seem to provide enough incentives to improve their practices. Bottom line is that reproducibility requires intrinsic motivation, a dedicated working attitude, and self-discipline, or otherwise can only be enforced with penalties and/or rewards. Indeed, it is well-recognized that the way in which we reward science should be changed. For example, the Declaration on Research Assessment (DORA) is a global initiative that proposes to change the evaluation of researchers and scholarly research output by funding agencies, academic institutions, and other parties53. At a broader level, UNESCO is also discussing the costs and benefits of open science and the incentives or motivations for open science54. Complementary, at the national level similar initiatives emerged. For example, the Dutch public knowledge institutes and research funders wrote a position paper that, among others, encourages Open Science55. The recently initiated Dutch OpenScience.nl organization will contribute to the implementation of some recommendations in this position paper. In addition, projects like Osiris aim to identify incentives for reproducibility by stakeholders, and embedding reproducibility in research design56. Due to such initiatives, we are slowly witnessing a change in the reward system, and we expect that this will largely contribute to more reproducible research. Stodden and co-workers suggested to have journals and/or professional societies discern a yearly award for (young) investigators for excellent reproducible practice52. Another proposal is to use scientific reproducibility as a litmus test for deciding between paper correction versus retraction57. Adoption of this guideline by journals would strongly support scientific reproducibility. ENCORE perfectly aligns with these suggestions and contributes to building the researcher’s scientific track record. An ENCORE project compendium can be shared through public repositories and assigned a DOI. We recently submitted the first ENCORE project to Zenodo42 and most of our future publications will be accompanied by an ENCORE project. In addition, we are preparing a publication describing specific challenges encountered when using ENCORE for a benchmark study for spatial transcriptomics deconvolution methods.
We consider ENCORE to be a step towards reproducible science, but it is not without several limitations and weaknesses. First, ENCORE is a compromise based on previous ways of working and, therefore, may not always fit the preferred way of working of an individual researcher. However, we believe ENCORE provides sufficient flexibility to accommodate most researchers and research practices. Second, the current sFSS Navigator has limited functionality, ways to present information and configuration options. We are in the process of improving the Navigator, which includes (i) configuration of panel locations (including the possibility to undock to a full window), (ii) Improve the presentation of figures and tables. (iii) improved possibilities to browse results and link results with code and data. (iv) Ensure proper formatting of text-based information (e.g., markdown, code) while ensuring that all relative hyperlinks work in the context of the Navigator. Third, a shortcoming of ENCORE is the lack of and clear and easy approach to specify explicit links between results, code, data, and concepts. Currently, such links are imposed by the sFSS structure, the paths in the code, and/or manually added links specified in the documentation. The documentation has an important function in gluing the project parts together, but it requires effort from the researcher to specify relations between parts of the project, and to maintain this information. ENCORE would benefit from improved integration approaches to enhance transparency and reproducibility. One possible approach is the use of JSON58 or YAML59 to annotate links between items in a project compendium as demonstrated by Spreckelsen and co-workers31. Fourth, another challenge that is only partially addressed by ENCORE concerns the preservation of the full computing environment. This environment is defined by (interdependencies of) the operating system, software tools, versions and dependencies, programming language libraries, etc. Gruning and co-workers proposed a software stack of interconnected technologies to preserve the computing environment33. This stack comprises (i) (Bio)Conda60,61 to provide virtual execution environments addressing software versions and dependencies, (ii) container platforms such as Docker62, Apptainer (formerly Singularity)63, and Podman64 to preserve other aspects of the runtime environment, and (iii) virtual machines using cloud systems or dedicated applications such as VMware, to overcome the dependencies on the operating system and hardware. We are currently investigating how to best approach this within the context of ENCORE but we are already actively using Anaconda and renv65 to manage Python libraries and R packages respectively. Reproducibility can further be improved by using scientific workflow systems, which have been developed to modularize and execute steps in computations. Many workflow systems are available, including Galaxy66, KNIME67, Snakemake68 and Nextflow69. These workflow systems improve reproducibility and help to maintain and share computational analyzes. They also allow incorporation of steps that otherwise would have been performed manually. Our group has used workflow systems in the past (e.g.70,71), but we decided not to make a workflow system an obligatory part of the ENCORE approach since we believe this may be too disruptive for some researchers. Yet, we encourage our group members to use a workflow system of choice. However, workflow systems are not the holy grail since these don’t solve the versioning problem of their (remote) components. In addition, the workflow system itself may become outdated, remote webservices may no longer be available, or older workflows may not run with newer versions of a workflow management system. Fifth, ENCORE projects can easily be shared but currently we do not have a good mechanism to remove parts that should not be shared such as sensitive (patient) data, controlled access data obtained from public repositories, or PDF copies of copyright protected scientific publications. Finally, ENCORE requires that all data used by the computations are within the sFSS structure. For large datasets this implies that sufficient storage space must be available on the computer that hosts the project, which can be a local (private) desktop computer, computer servers of the research institute, or remote (cloud) compute systems (e.g., as provided by Amazon Web Services; Fig. 3). If data storage within the sFSS is not possible then documentation and/or software should be available to retrieve the data from another persistent location and to ensure that this step does not break reproducibility.
Over the past two decades, an increasing number of biomedical researchers have become involved in computational research. Many of these researchers have never been formally trained in scientific computing and software engineering (e.g., design, programming, documentation)72,73, software version control25,27, the use of high-performance computing infrastructures, the use of Unix/Linux which is still the major platform for scientific computing, algorithm design, the use of (Jupyter, R) notebooks74, etc. Lack of such skills may negatively affect reliability and transparency of software and, consequently, reproducibility. For example, software may be poorly designed and documented, making it difficult to understand, use, modify, and debug. One resulting problem is that we have no way of knowing whether the code being used to generate the computational results is doing what the researchers think it is doing. This is one reason why ENCORE proposes to start organizing and documenting from the start of a project, since this increases the chance that conceptual errors or software bugs are detected in an early stage by the researchers or their supervisors. Software engineering is a discipline in its own and includes the design, implementation, documentation, testing and deployment of software. Following best practices for scripting, functional programming, or object-oriented programming may significantly improve the quality of the code but requires training and experience. The use of integrated development environments, automated quality checks, and (unit) testing would also help to improve software75. Furthermore, Large Language Models will increasingly play a role in software development, testing, and documentation76,77. Often, software documentation leaves much to desire. In a recent report, it was concluded that researchers are generally not aware for whom they write documentation and what documentation is required73. Currently, ENCORE does not provide specific instructions for coding style (e.g., PEP 8 for Python and tidyverse for R78,79) and documentation design because it is probably more effective to train scientists in the art of software engineering. Instead, general guidelines are provided in a README file (Supplementary Method 3). Awareness of guidelines (e.g.80) and tools to (automatically) generate documentation such as Sphinx81 for Python, and r2readthedocs82 and roxygen283 for R, will also help to improve reproducibility. We used Sphinx for the documentation of the sFSS Navigator42. Another useful resource is the software management plan developed by the Netherlands eScience Center and the Dutch Research Council (NWO)84. In general, appropriate training on reproducibility approaches could already significantly improve the current situation and will at least create awareness of the tremendous amount of literature about many aspects of sound scientific computing practices20,39,85,86,87,88. In addition, senior researchers should strongly promote reproducibility and support, explain, and instruct junior researchers.
To facilitate and improve reproducibility throughout the complete research lifecycle (Fig. 1), numerous guidelines, policies, and standards have been developed89 to guide and facilitate the detailed documentation of all steps. Many reporting guidelines provide structured tools specifying the minimum information required for specific study types and largely contribute to the transparency, understanding, and reproducibility of a study90. Examples include guidelines for clinical trials (CONSORT)91, diagnostic accuracy studies (STARD)92, and observational studies (STROBE)93. Virtually all these minimum information standards and reporting guidelines require the specification of statistical and computational methods that were used in a study. However, precise requirements to specify such methods are often lacking. In addition, an increasing number of scientific journals have their own guidelines. One example is the Nature Reporting Summary94 that partially relies on existing reporting guidelines and FAIR. Nature also has a specific software and algorithm policy with requirements about availability (e.g., using GitHub or Zenodo), use of an open-source license10, and code review95. Nature Computational Science and several other Nature journals have adopted Code Ocean, which is a cloud-based platform to share executable code to enable review96,97. The PLOS Computational Biology journal requires that editors and reviewers can access the software, reproduce the results, and run the software on a deposited dataset with provided control parameters98. The Science journal TOP guidelines require data and computer code to be available99. Interestingly, a study published in 2018 showed that despite these guidelines, many computational studies were not reproducible17. It has also been suggested to rethink the concept of a “journal” as a community-driven information repository containing, among others, the data and software, to enable reproducibility, reuse, and comparisons100. In this scenario, academic publishers would have a key role in stimulating (standard-based) approaches to research dissemination. We believe that such an effort should be a joint undertaking of research communities and publishers aiming to improve reproducibility. Computational projects require their own specific guidelines and standards to guarantee transparency and reproducibility. This was also recognized by the artificial intelligence community, which started initiatives to develop specific AI-oriented guidelines101,102,103. To the best of our knowledge, there are not many (practical) reporting guidelines nor standards available for computational research that are routinely used in practice. Nevertheless, there are various initiatives to improve this situation. For example, the use of directory structures to organize research projects has been proposed in the past, However, in general these initiatives do not provide specific templates or are limited to a specific programming language20,36,104. In particular, the approach presented by (Marwick, 2018) is useful for R-based projects and uses the ‘rrtools’ package to setup a project compendium suitable for writing a reproducible manuscript. It supports Quarto (an open-source scientific and technical publishing system), Docker, package versioning using renv, and integration with GitHub Actions. ENCORE is inspired by the standardized file system structure that was proposed by Spreckelsen and co-workers31. Their file system structure comprises four top-level directories denoted as categories (Experimental data, Simulations, Data analysis, and Publication). Each of these categories contains subdirectories to hold specific projects in which other subdirectories may exist. This implies that, in practice, each of the four top-level directories will contain subdirectories and files related to multiple projects. The subdirectories and files are manually annotated and connected using YAML headers (key-value pairs) in README Markdown files, which allows, for example, to trace simulations and data belonging to a specific project. The ENCORE sFSS is project-oriented, which facilitates sharing with peers without first having to reassemble a project. At the same time this reduces the need to manually annotate and define relationships, since these are implied by the sFSS structure. Nevertheless, ENCORE would also benefit from integration approaches such as YAML to improve transparency and reproducibility. However, ENCORE does not yet require the use of YAML since it may require too much effort from researchers to specify and maintain. Like ENCORE, the approach of Spreckelsen et al. has a strong focus on transparently organizing and linking data, code, and publications to support reproducibility. However, there are no further requirements except that one should be able to re-run code from within their file system structure. In addition, there are no requirements by design for further user and code documentation and the use of GitHub. Compared to the approach of Spreckelsen et al., ENCORE also aims to translate published guidelines into specific instructions and templates in, for example, the README Markdown files to guide researchers in making their computational work reproducible. This is supported by providing our sFSS as a template together with predefined files. In contrast to the file system structure proposed by Spreckelsen, the ENCORE sFSS is much more detailed. ENCORE goes beyond a structured file system and through regular internal and external evaluations we aim to gradually improve the ENCORE approach to improve reproducibility. Other examples of guidelines and standards for computational projects include the application of the FAIR principles to software105,106, the ICERM implementation and archiving criteria for software52, the Adaptive Immune Receptor Repertoire (AIRR) software guidelines107, the Software Ontology to describe software used to store, manage and analyze data108, and the EDAM ontology to describe bioinformatics operations109. For simulation-based research, there are initiatives like the Minimum Information About a Simulation Experiment (MIASE) guidelines110, the corresponding simulation experiment description markup language (SED-ML)111, COMBINE/OMEX to share and reproduce modeling projects112, and a range of others113. For the further development of ENCORE, we will need to consider which of these standards are relevant for ENCORE and how to incorporate them in the ENCORE approach. This may require the development of ENCORE specifications for different types of computational projects by different specialized working groups. However, the main challenge we see is the development of software tools to support and use ontologies and standards in the context of ENCORE without introducing much overhead while providing clear benefit.
Currently, the focus of ENCORE is on human readability and not machine readability since most documentation will be provided in any of the common file types (e.g., plain text, Markdown, Word, PowerPoint and PDF), which has obvious advantages for the researcher. However, the additional use of text-based standards such as JSON, YAML, and RDF114, would allow to (partially) document projects in a machine readable manner and provide several benefits. For example, one might more easily search for specific information within a project compendium, specify links between items in a compendium, and to evaluate compliance of the project organization and documentation with the ENCORE requirements. To advance ENCORE towards a machine-readable compendium, one approach would be to associate subdirectories and/or files with JSON annotation (meta-data) and, where possible, to provide part of the documentation directly in a JSON file. This approach would still allow the researcher to use the file type of choice. Alternatively, one may integrate YAML annotation directly into Markdown files as proposed by Spreckelsen115. Finally, realizing that these machine-readable formats can be rendered by front-end GUI applications (e.g., as text fields for users to enter), yet another approach would be the development of interactive tools for users to set up and populate the documentation in their projects. This would remove much of the burden from the researcher to specify and maintain the JSON/YAML/RDF annotation throughout a project.
To promote reproducibility practices using ENCORE, automating specific steps becomes invaluable in lightening the load on researchers. Leveraging AI-based tools can play a pivotal role in tasks such as code documentation or testing, summarizing project meetings, ensuring compliance with ENCORE specifications, and using machine-readable files effectively. These automation efforts streamline workflows, enhance accuracy, and ultimately contribute to the robustness of research endeavors. We have created a separate GitHub repository (https://github.com/EDS-Bioinformatics-Laboratory/ENCORE-AUTOMATION) to which we will gradually add tools to automate specific tasks. Currently, we provide scripts to automatically setup an ENCORE project. In addition, we provide a script that generates an ENCORE template for B-cell/T-cell repertoire sequencing experiments116, which is prefilled with code and documentation. An example of such generated template is found on GitHub (https://github.com/EDS-Bioinformatics-Laboratory/ENCORE-AIRRseq-TEMPLATE). A similar template is currently being developed for lipidomics analyzes117.
Methods
ENCORE comprises a standardized file system structure (sFSS), pre-defined (Markdown) files for documentation, a GitHub repository for software version control, a HTML-based sFSS browser (sFSS Navigator), and ENCORE documentation to initiate a self-contained project compendium and support the researcher to use ENCORE in a consistent manner. The sFSS template can be retrieved from the ENCORE GitHub repository (https://github.com/EDS-Bioinformatics-Laboratory/ENCORE)30. In principle, ENCORE can be used for any type of (research) project and is not domain-specific.
The ENCORE approach was driven by the following main requirements:
-
1.
Consist of a single self-contained project compendium. The computational project should be organized and available as a self-contained and integrated compendium of data, code, results, and (conceptual) documentation, stored at a single location. It should also be easily transferable to other researchers or reviewers without breaking its internal consistency.
-
2.
Facilitate transparency and documentation. ENCORE should facilitate transparency and a deep understanding (e.g., addressing why specific methods were selected and how these were applied) of the project through its standardized structure and documentation of concepts, methodology, data, code, and results.
-
3.
Enable reproducibility. The project compendium should enable an (independent) peer to autonomously execute and understand the computational techniques and recreate the (published) outcomes.
-
4.
Adhere to proposed guidelines. ENCORE should follow published guidelines for computational reproducibility as much as possible (e.g.19,32,34,36).
-
5.
Enable version control. ENCORE should allow version control of code and code documentation.
-
6.
Facilitate harmonization. The ENCORE approach itself should be standardized and well-documented such that it can easily be adopted by any researcher. This allows harmonization within research groups, enabling further joint development of best practices within the ENCORE framework. Moreover, harmonization also facilitates checking transparency and reproducibility prior to publication by direct colleagues.
-
7.
Provide a generic approach. ENCORE should be agnostic to the type of computational project (e.g., statistical analysis, mathematical modeling), data, programming language, and ICT infrastructure (e.g., operating system and computer hardware). ENCORE should make use of a software versioning system but otherwise should not rely on tools for project management, data processing, etc. See also Supplementary Method 4.
-
8.
Allow adaptation to different styles of working. ENCORE should leave sufficient flexibility to accommodate different styles of working. The underlying sFSS should be accessible from any software tool the researcher might be using.
In addition to these requirements, we defined four principles to provide practical guidance for researchers (Supplementary Method 1, Section 2).
The sFSS in context
The sFSS is the project’s entry point (Fig. 4) and, therefore, should be self-contained, which is the responsibility of the project team. The software developed in the project, and the external software documentation is additionally managed through Git and the project’s GitHub repository to enable version control and joint development. The principled decision to only make code and code documentation available on GitHub and exclude other project components, such as data and results, is motivated by the requirement to create a self-contained sFSS project compendium while GitHub is not a universal storage platform. In addition, the size of the data and results may be too large to host on GitHub. Consequently, in practice, an sFSS contains only the software version that will be shared, while the GitHub repository may also contain older or alternative versions not required to reproduce the computational results. The complete project compendium can be shared with other researchers or reviewers or submitted to public repositories such as Zenodo for archiving, in which case a DOI can be assigned. Providing third parties access to the GitHub repository is optional.
The green box denotes the Project Compendium (sFSS) with part of the directory structure shown. The sFSS is the central point of entry for a project and is initially cloned from the ENCORE template GitHub repository when starting a new project. The project team is responsible for the organization and documentation of the project. Only the code and code documentation within the project compendium are synchronized to a project specific GitHub repository. An sFSS project compendium can be shared with a compendium recipient. The compendium recipient starts exploring the project by opening Navigate.html in a web browser.
Instantiation of a new ENCORE-based project
ENCORE enables several approaches to set up a new project. The approach used for most research projects is that the project team first clones and initializes the sFSS template from the ENCORE GitHub repository (Fig. 4) by the project team. Subsequently, one creates a new project specific GitHub repository and connects this to the Processing directory (Fig. 5) of the project to ensure that only code and code documentation will be synchronized with the GitHub repository. Part of the pre-defined files are provided in different formats (e.g., txt, tex, docx) for the project team to make a quick start. They select the format of preference and, before sharing, remove the files (e.g., Step-by-Step ENCORE guide) that are not relevant for the compendium recipient with whom the project will be shared in the future (Supplementary Method 2). Finally, one starts documenting the project. These steps are described in detail in the Step-by-Step ENCORE guide (Supplementary Method 1) and takes less than 30 minutes to complete. Another option is to use a pre-defined script to automatically instantiate a new project (available from https://github.com/EDS-Bioinformatics-Laboratory/ENCORE_AUTOMATION)118. For specific cases one may write dedicated scripts to setup a new project (see Discussion).
The main component comprises the sFSS template (1) that organizes all parts of the project. ‘Project’ corresponds to the root directory of the template. The blocks represent project-dependent sub-directories (Fig. 4). Project documentation resides in (pre-defined) files (2) that are found in all subdirectories of the sFSS template. The pre-defined files contain instructions about the minimum information that needs to be provided in terms of documentation for the different parts of a project. Each project is complemented with a GitHub repository (3) for version control of the code and documentation in the ‘Processing’ (sub-)directories. The sFSS Navigator (4) allows (end) users to browse the main contents of the project. The external ENCORE user documentation (5) provides instructions for new users on how to instantiate a new project.
The ENCORE components
ENCORE comprises five main components (Fig. 5) to structure, integrate, control code and documentation versions, and to improve transparency and reproducibility of a project.
Component 1. The standardized File System Structure (sFSS) template
The sFSS provides a standardized, yet flexible, template to organize conceptual information, data, code, documentation, and (intermediate) results (e.g., tables, figures, text files). In essence, it is a standardized directory structure containing pre-defined files, which can be used with any operating system (Fig. 6) and can be used with any data analysis software that reads from and writes to a file system. Conceptual information includes any information considered relevant to correctly setup the computations, but also information that helps peers to understand the (background of the) project and information that documents why specific choices were made. This includes information about the research question, experimental design, samples used, wet-lab experiments, description of computational (for example, statistical or mathematical modeling) approaches, interpretation of results, relevant literature, presentations about the project, and summaries from (email) discussions during the project. Files within the sFSS may exist in different versions if, for example, new data were generated, code was modified, or figures were updated based on different settings for the computations. The information within the sFSS is implicitly and explicitly integrated. Implicit links correspond to relations that are imposed due to the hierarchical structure of the sFSS. Explicit links are made in the (code) documentation, for example, by linking a particular computation to a specific subset of the data. The sFSS allows a certain degree of flexibility to accommodate different types of projects or different ways of working. For example, data can be organized at different levels in the sFSS (Fig. 5). This allows, for example, to organize the data directly within the subdirectory of a specific computation, if that data is not used by other computations. Similarly, if a project involves the (pre)processing of data, then the outcome of these steps may either be considered as a ‘Result’ in a NameOfComputation directory, or as ‘Processed data’ in a Data directory (Fig. 6). The ‘LabJournal’ can be copied to other directories to accommodate more specific documentation. Markdown README files for project documentation may be replaced with other file types (e.g. HTML, LaTeX) if one prefers. Pre-defined sFSS directories and files that are not used can be removed. Detailed information about the specific structure of the sFSS directory structure can be found in Supplemenatary Method 2.
Standardized directory structure of the sFSS containing pre-defined files (gold), which include README files (in Markdown format) that provide a documentation template and instructions. Note that the pre-defined files in the ‘Data’ directories (orange) and the ‘0_SoftwareEnvironment’ subdirectories are only shown once. The names of the directories ‘NameOfDataset_1’ and ‘NameOfComputation_1’ are placeholders and should be replaced with more descriptive names. These directories can be replicated if multiple datasets are used or if different computation procedures are performed. Subdirectories shown in light blue are under version control using Git/GitHub. The ‘0_’ prefix ensures that the corresponding files/directories are always on top of the file list when using lexicographic ordering. The README.md in ‘Processing’ is the default GitHub repository README file and therefore does not have the ‘0_’ prefix.
Component 2. Pre-defined files
The sFSS contains many pre-defined files that should be used directly from the start of the project and maintained throughout the project (Fig. 6). Most files are in Markdown format, which was chosen because it is the default format for the README file of a GitHub repository. Markdown is a markup language, which enables adding formatting elements to plain text119 and requires a compatible editor. However, if preferred, these may be replaced by other file formats (e.g., docx, HTML, tex). In the sFSS root directory, the 0_PROJECT.md file should provide basic project details, including the project name, start date, a short description, and project team. In addition, the 0_GETTINGSTARTED template (docx, HTML, tex, txt) should describe the most important aspects of the project and should provide links to the relevant sub-directories and files. This final template is then saved as an HTML file and together with 0_PROJECT.md used by the sFSS Navigator (see below). The LabJournal template (docx, md, tex, txt) in the ProjectDocumentation directory should contain general project documentation, including but not limited to an explanation of the project’s background and concepts, computational approaches, summaries of project discussions, new research ideas, and to-do lists. Preferably, the lab journal should contain pointers to relevant sub-directories and files whenever needed (which is easy with Markdown). Alternatively, one may maintain multiple lab journals in different directories containing documentation for specific parts of the project. The lab journal is important for the scientific legacy of the research group by ensuring that others can replicate what the original researcher(s) has done. We decided to deviate from standard practice and to also use the lab journal to record new ideas, provide summaries of (email) discussions, and to-do lists, since it is important to have a record of these for the supervision of the projects and for follow-up projects. Consequently, not all information in the lab journal can be shared with others (see Discussion). Each directory also contains a Markdown README file that is specific for the directory in which it is located. In general, these README files contain an explanation of the information found in that specific directory, instructions, and a documentation template specifying the minimum required documentation that a researcher should provide (Supplemenatary Method 3). The instructions and templates are basically a translation of a selection of published guidelines to enhance the reproducibility of computational projects. This is an essential part of ENCORE since following the instructions and templates will likely improve the reproducibility of the project while preventing the researcher from reading the many publications about computational reproducibility to deduce what should be documented and why. The instructions and templates also support the internal consistency of the sFSS and promote consistency between different ENCORE projects within the same research group. One decision to be made by the project team is how to distribute the documentation over the various README files and the lab journal(s). However, as a rule, one should document any project file (code, data, results) in the directories in which these are located. The lab journal can then be used for more general documentation.
Component 3. The GitHub repository
ENCORE utilizes Git and GitHub for version control of software and its documentation, and to collaborate on software development. Since the sFSS project compendium contains all the information of a project, it is not necessary to also share the GitHub repository. However, the project team may still share this repository or make it public in case of joint code development or if access to previous code versions is requested or required. To ensure that only code, (Jupyter) notebooks, software settings, and documentation is managed by Git, the project team needs to configure the so-called ‘.ignore’ file of which templates are provided in the sFSS (Supplemenatary Method 1).
Component 4. The sFSS navigator
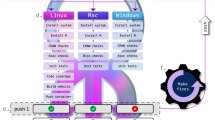
At later stages of a project, the sFSS may contain a large amount of information potentially making it difficult for the compendium recipient to determine the best point of entry. Therefore, we developed the sFSS Navigator to provide a first overview of the project. It also provides a convenient tool to, for example, browse and show results while the project is still active. The project team uses the 0_GETTINGSTARTED file to provide the compendium recipient pointers to directories and files that contain the most relevant information such as the main results and the data and code that were used to generate these. The sFSS Navigator itself was developed following the ENCORE approach, and the project compendium can be found in Zenodo42. The Navigator is a Python program, which scans the sFSS and generates a web page (Navigate.html) that can be opened in any web browser. The generated web page consists of four panels (Fig. 7). The content of the panels is configured by the project team to guide peers to the important parts of the project. A configuration file (Navigation.conf) allows to specify which subdirectories and files to show in the expandable directory tree (Fig. 7a). In addition to the Python code (Navigate.py), executables for Windows, macOS (for both Intel- and silicon-based chips) and shell scripts for Linux/Unix using Python v3 are provided to ensure the Navigator can be used if the Python interpreter is not installed.
a Expandable sFSS directory tree and link to the project’s GitHub repository. The project team can configure, which directories and files to show. b Content of a selected file. In this example, the panel shows the content of the default GitHub Markdown README file. c General project description, contact person, and collaborators (0_PROJECT.md). d Getting started explains the project and includes links to the various files and directories in the sFSS (0_GettingStarted.html).
Component 5. User documentation
ENCORE is complemented with extensive documentation to guide the user in setting up a new project and maintaining documentation throughout the project. In addition to the documentation already present in the pre-defined files, we have created a comprehensive ‘Step-by-Step ENCORE Guide’ (Supplemenatary Method 1). This guide offers a brief introduction to the ENCORE principles and components and provides a recipe for instantiating an sFSS for a specific project, a corresponding GitHub repository, and the sFSS Navigator. It also includes a basic introduction to Git and GitHub to troubleshoot problems.
General ENCORE guidelines
The initialization of an ENCORE-based project is straightforward and does not take much time. However, it is important to keep working according to the provided instructions and to keep the documentation continuously up to date. Recollecting (from memory) all important details at the end of a project is virtually impossible. Moreover, this will take significantly more time than incrementally adding documentation during the project. While organizing and documenting a project, one should assume that at some stage the project is going to be shared with a peer who is initially unfamiliar with the project, the computational concepts, the type of data, and the programming language. Although it may be tempting to document all aspects of the project in a single document eventually evolving into a manuscript to be submitted for peer review, we advise not to do so for several reasons. First, it breaks the connection between the documentation and the files (e.g., code or data) being documented. Second, the level of documentation in an sFSS is likely to be much more detailed than what is provided in a typical research paper. Even the manuscript’s supplementary information may not provide the same level of detail. Third, since ENCORE is used right from the start of a project, it is likely that not all parts of the project end up in a publication. For example, not all (intermediate) results will become part of a publication. In addition, documenting specific approaches that failed or were discontinued may be equally important. Fourth, keeping the documentation modular and in the directories where it belongs helps in its maintenance. Once the manuscript will be written those parts of the documentation that are needed can then easily be incorporated. We also recommend that the overall sFSS structure, names of directories (with exceptions mentioned earlier), and names of pre-defined files are not changed. This holds in particular for research groups that embrace this approach to achieve harmonization. Additional directories may be added if this does not effectively change the overall sFSS structure. For example, within the Results directory, one may create separate sub-directories for figures and tables, but one should not move the Results directory to the root of the sFSS. As mentioned earlier, unused directories and files should be removed. Since an sFSS should be self-contained, any directory path used within the code or in any of the pre-defined files should be relative to the top-level directory.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Code availability
The ENCORE template described in this manuscript is available as a GitHub release (https://github.com/EDS-Bioinformatics-Laboratory/ENCORE/releases/tag/V4.1.1) (https://doi.org/10.5281/zenodo.12938252) and licensed under CC BY NC SA 4.0. The ENCORE_AUTOMATION GitHub repository contains the scripts referred to from this manuscript (https://github.com/EDS-Bioinformatics-Laboratory/ENCORE_AUTOMATION) which are also available from https://doi.org/10.5281/zenodo.12955697 and are Licenced under GNU GPL V2. The sFSS Navigator is available from Zenodo (https://doi.org/10.5281/zenodo.7985655) or its GitHub repository (https://github.com/EDS-Bioinformatics-Laboratory/FSS-Navigator) and is licensed under GNU GPL V2.
References
Stupple, A., Singerman, D. & Celi, L. A. The reproducibility crisis in the age of digital medicine. NPJ Digit Med 2, 2 (2019).
Ioannidis, J. P. Why most published research findings are false. PLoS Med 2, e124 (2005).
Fidler F., Wilcox J. Reproducibility of scientific results. the Stanford Encyclopedia of Philosophy. https://plato.stanford.edu/archives/sum2021/entries/scientific-reproducibility (2021).
Diaba-Nuhoho, P. & Amponsah-Offeh, M. Reproducibility and research integrity: the role of scientists and institutions. BMC Res Notes 14, 451 (2021).
Turkyilmaz-van der Velden, Y., Dintzner, N. & Teperek, M. Reproducibility starts from you today. Patterns (N. Y) 1, 100099 (2020).
Merino Tejero, E. et al. Multiscale modeling of germinal center recapitulates the temporal transition from memory B cells to plasma cells differentiation as regulated by antigen affinity-based Tfh cell help. Front Immunol. 11, 620716 (2020).
Lashgari, D. et al. From affinity selection to kinetic selection in Germinal Centre modelling. PLoS Comput Biol. 18, e1010168 (2022).
van der Heyden, M. A. G. & van Veen, T. A. B. Gold open access: the best of both worlds. Neth. Heart J. 26, 3–4 (2018).
Pulverer, B. Open access-or open science? EMBO J. 37, e101215 (2018).
Open Source Initiative. OSI approved licenses. https://opensource.org/licenses (2023).
Garijo, D. et al. Nine best practices for research software registries and repositories. PeerJ Comput Sci. 8, e1023 (2022).
Rigden, D. J. & Fernandez, X. M. The 2023 nucleic acids research database Issue and the online molecular biology database collection. Nucleic acids Res. 51, D1–D8 (2023).
Figshare. Repository to make research outputs available in a citable, shareable and discoverable manner. https://figshare.com (2023).
Zenodo. E. U. open research repository. https://doi.org/10.25495/25497gxk-rd25471 (2023).
Papin, J. A., Mac Gabhann, F., Sauro, H. M., Nickerson, D. & Rampadarath, A. Improving reproducibility in computational biology research. PLoS Computational Biol. 16, e1007881 (2020).
Tiwari, K. et al. Reproducibility in systems biology modelling. Mol. Syst. Biol. 17, e9982 (2021).
Stodden, V., Seiler, J. & Ma, Z. An empirical analysis of journal policy effectiveness for computational reproducibility. Proc. Natl Acad. Sci. USA 115, 2584–2589 (2018).
Mendes, P. Reproducible Research Using Biomodels. Bull. Math. Biol. 80, 3081–3087 (2018).
Sandve, G. K., Nekrutenko, A., Taylor, J. & Hovig, E. Ten simple rules for reproducible computational research. PLoS Computational Biol. 9, e1003285 (2013).
Wilson, G. et al. Good enough practices in scientific computing. PLoS Computational Biol. 13, e1005510 (2017).
Ziemann, M., Poulain, P. & Bora, A. The five pillars of computational reproducibility: bioinformatics and beyond. Brief. Bioinforma. 24, bbad375 (2023).
Hunter-Zinck, H., de Siqueira, A. F., Vasquez, V. N., Barnes, R. & Martinez, C. C. Ten simple rules on writing clean and reliable open-source scientific software. PLoS Comput Biol. 17, e1009481 (2021).
Deardorff, A. Assessing the impact of introductory programming workshops on the computational reproducibility of biomedical workflows. PloS one 15, e0230697 (2020).
Margan D., Candrlic S. The Success Of Open Source Software: A Review. 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO); Opatija 1463-1468 (2015).
Blischak, J. D., Davenport, E. R. & Wilson, G. A quick introduction to version control with Git and GitHub. PLoS Computational Biol. 12, e1004668 (2016).
Ram, K. Git can facilitate greater reproducibility and increased transparency in science. Source Code Biol. Med 8, 7 (2013).
Perez-Riverol, Y. et al. Ten simple rules for taking advantage of Git and GitHub. PLoS Computational Biol. 12, e1004947 (2016).
Wilkinson, M. D. et al. The FAIR guiding principles for scientific data management and stewardship. Sci. Data 3, 160018 (2016).
Di Cosmo, R. Archiving and referencing source code with software heritage. Math. Softw. - ICMS 2020 12097, 362–373 (2020).
van Kampen A. H. C., et al. ENCORE: a practical implementation to improve reproducibility and transparency of computational research. ENCORE https://zenodo.org/records/12938252, (2024).
Spreckelsen, F. et al. Guidelines for a standardized filesystem layout for scientific data. Data 5, 43 (2020).
Brito, J. J. et al. Recommendations to enhance rigor and reproducibility in biomedical research. Gigascience 9, giaa056 (2020).
Gruning, B. et al. Practical computational reproducibility in the life sciences. Cell Syst. 6, 631–635 (2018).
Lee, B. D. Ten simple rules for documenting scientific software. PLoS Computational Biol. 14, e1006561 (2018).
Markowetz, F. Five selfish reasons to work reproducibly. Genome Biol. 16, 274 (2015).
Noble, W. S. A quick guide to organizing computational biology projects. PLoS Computational Biol. 5, e1000424 (2009).
Stodden, V. & Miguez, S. Best practices for computational science: software infrastructure and environments for reproducible and extensible research. J. Open Res. Softw. 2, e21 (2014).
Taschuk, M. & Wilson, G. Ten simple rules for making research software more robust. PLoS Computational Biol. 13, e1005412 (2017).
Wilson, G. et al. Best practices for scientific computing. PLoS Biol. 12, e1001745 (2014).
Stodden, V. Reproducing statistical results. Annu. Rev. Stat. Its Application 2, 1–19 (2015).
Types of GitHub accounts. https://docs.github.com/en/get-started/learning-about-github/types-of-github-accounts (2023).
Van Kampen, A. H. C., Jongejan, A. & Mahamune, U. The standardized file system structure (FSS) navigator repository. FSS-Navigator https://doi.org/10.5281/zenodo.7985655 (2023).
National Academies of Sciences E, Medicine. Reproducibility and Replicability in Science. (The National Academies Press, 2019).
Patil, P., Peng, R. D. & Leek, J. T. A visual tool for defining reproducibility and replicability. Nat. Hum. Behav. 3, 650–652 (2019).
Milkowski, M., Hensel, W. M. & Hohol, M. Replicability or reproducibility? on the replication crisis in computational neuroscience and sharing only relevant detail. J. Comput Neurosci. 45, 163–172 (2018).
Plesser, H. E. Reproducibility vs. replicability: a brief history of a confused terminology. Front Neuroinform 11, 76 (2017).
Lyon, L. Transparency: the emerging third dimension of open science and open data. Lib. Q. 25, 153–171 (2016).
Anderson, J. A., Eijkholt, M. & Illes, J. Ethical reproducibility: towards transparent reporting in biomedical research. Nat. methods 10, 843–845 (2013).
Kulikowski, C. & Maojo, V. M. COVID-19 pandemic and artificial intelligence: challenges of ethical bias and trustworthy reliable reproducibility? BMJ Health Care Inf. 28, e100438 (2021).
ICMJE. International Committee of Medical Journal Editors. Defining the role of authors and contributors. https://www.icmje.org/recommendations/browse/roles-and-responsibilities/defining-the-role-of-authors-and-contributors.html (2023).
Editorial. Supporting computational reproducibility through code review. Nat. Hum. Behav. 5, 965–966 (2021).
Stodden V., et al. Setting the default to reproducible reproducibility in computational and experimental mathematics. Preprint at https://icerm.brown.edu/topical_workshops/tw12-5-rcem/icerm_report.pdf (2012).
DORA. The declaration on research assessment. https://sfdora.org (2023).
Implementation of the UNESCO recommendation on open science. working group on open science funding and incentives, https://unesdoc.unesco.org/ark:/48223/pf0000383806 (2022).
VSNU, NFU, KNAW, NWO, ZonMw. Room for everyone’s talent: towards a new balance in the recognition and rewards for academics. https://recognitionrewards.nl/ (2019).
Osiris. Open Science to Increase Reproducibility in Science. https://osiris4r.eu/ (2024).
Moseley, H. In the AI science boom, beware: your results are only as good as your data. Nature https://doi.org/10.1038/d41586-024-00306-2 (2024).
JSON. JavaScript Object Notation. https://www.json.org (2024).
YAML Ain’t Markup Language. https://yaml.org (2023).
Gruning, B. et al. Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. methods 15, 475–476 (2018).
Anaconda Software Distribution. https://docs.anaconda.com (2023).
Nust, D. et al. Ten simple rules for writing dockerfiles for reproducible data science. PLoS computational Biol. 16, e1008316 (2020).
Kurtzer, G. M., Sochat, V. & Bauer, M. W. Singularity: Scientific containers for mobility of compute. PloS one 12, e0177459 (2017).
Podman. A tool designed to find, run, build, share and deploy applications using Open Containers Initiative (OCI). https://podman.io (2024).
Ushey K., Wickham H. renv: Project environments. https://rstudio.github.io/renv (2024).
Galaxy, C. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic acids Res. 50, W345–W351 (2022).
Berthold M. R., et al. KNIME: The Konstanz Information Miner. In: Data Analysis, Machine Learning and Applications (eds Preisach C., Burkhardt H., Schmidt-Thieme L., Decker R.). Springer Berlin Heidelberg (2008).
Molder, F. et al. Sustainable data analysis with Snakemake. F1000Res 10, 33 (2021).
Di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35, 316–319 (2017).
Madougou, S. et al. Provenance for distributed biomedical workflow execution. Stud. health Technol. Inform. 175, 91–100 (2012).
Boekel, J. et al. Multi-omic data analysis using Galaxy. Nat. Biotechnol. 33, 137–139 (2015).
Martin R. C. Clean Code: A Handbook of Agile Software Craftsmanship. Pearson (2008).
Hermann, S. & Fehr, J. Documenting research software in engineering science. Sci. Rep. 12, 6567 (2022).
Rule, A. et al. Ten simple rules for writing and sharing computational analyses in Jupyter Notebooks. PLoS computational Biol. 15, e1007007 (2019).
Ebert, C., Bajaj, D. & Weyrich, M. Testing Software Systems. IEEE Softw. 39, 8–17 (2022).
Wang, J. et al. Software testing with large language models: survey, landscape, and vision. IEEE Trans. Softw. Eng. 50, 911–936 (2024).
Copilot G. AI coding assistant. https://github.com/features/copilot (2024).
Tidyverse Style Guide. https://style.tidyverse.org (2024).
Python Style Guide. https://www.python.org/doc/essays/styleguide/ (2024).
Karimzadeh, M. & Hoffman, M. M. Top considerations for creating bioinformatics software documentation. Brief. Bioinforma. 19, 693–699 (2018).
Brandl G. Sphinx documentation. https://www.sphinx-doc.org (2021).
r2readthedocs. Convert R package documentation to a ‘readthedocs’ website, https://github.com/ropenscilabs/r2readthedocs (2023).
Roxygen2. Dynamic documentation system. https://cran.r-project.org/web/packages/roxygen2 (2023).
Martinez-Ortiz C., et al. Practical guide to software management plans (1.1). https://doi.org/10.5281/zenodo.7589725 (2023).
Toelch, U. & Ostwald, D. Digital open science-Teaching digital tools for reproducible and transparent research. PLoS Biol. 16, e2006022 (2018).
Carey, M. A. & Papin, J. A. Ten simple rules for biologists learning to program. PLoS computational Biol. 14, e1005871 (2018).
Larcombe L., et al. ELIXIR-UK role in bioinformatics training at the national level and across ELIXIR. F1000Res 6, (2017).
Lapp, Z. et al. Developing and deploying an integrated workshop curriculum teaching computational skills for reproducible research. J. Open Source Educ. 5, 144 (2022).
FAIRsharing.org. A curated, informative and educational resource on data and metadata standards, inter-related to databases and data policies. https://fairsharing.org (2023).
EQUATOR Network. What is a reporting guideline? https://www.equator-network.org/about-us/what-is-a-reporting-guideline (2023).
Schulz, K. F., Altman, D. G., Moher, D. & Group, C. CONSORT 2010 statement: updated guidelines for reporting parallel group randomized trials. Ann. Intern Med 152, 726–732 (2010).
Bossuyt, P. M. et al. STARD 2015: an updated list of essential items for reporting diagnostic accuracy studies. BMJ 351, h5527 (2015).
von Elm, E. et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Ann. Intern Med 147, 573–577 (2007).
Nature. Reporting Summary. https://www.nature.com/documents/nr-reporting-summary-flat.pdf (2023).
Nature. Availability and peer review of computer code and algorithm. https://www.nature.com/nature-portfolio/editorial-policies/reporting-standards#availability-of-computer-code (2023).
Editorial. Seamless sharing and peer review of code. Nat. Comput Sci. 2, 773 (2022).
Editorial. Easing the burden of code review. Nat. methods 15, 641 (2018).
PLOS Computational Biology. Material, Software, and Code Sharing. https://journals.plos.org/ploscompbiol/s/materials-software-and-code-sharing (2023).
Science. Research Standards. Transparency and Openness Promotion (TOP) guidelines. https://www.science.org/content/page/science-journals-editorial-policies#TOP-guidelines (2023).
Gavaghan, D. Problems with the current approach to the dissemination of computational science research and its implications for research integrity. Bull. Math. Biol. 80, 3088–3094 (2018).
Ibrahim, H. et al. Reporting guidelines for clinical trials of artificial intelligence interventions: the SPIRIT-AI and CONSORT-AI guidelines. Trials 22, 11 (2021).
Ibrahim, H., Liu, X. & Denniston, A. K. Reporting guidelines for artificial intelligence in healthcare research. Clin. Exp. Ophthalmol. 49, 470–476 (2021).
Haibe-Kains, B. et al. Transparency and reproducibility in artificial intelligence. Nature 586, E14–E16 (2020).
Marwick, B., Boettiger, C. & Mullen, L. Packaging data analytical work reproducibly using R (and Friends). Am. Statistician 72, 80–88 (2018).
Barker, M. et al. Introducing the FAIR principles for research software. Sci. Data 9, 622 (2022).
Five Recommendations for FAIR Software. https://fair-software.eu (2023).
AIRR Software Guidelines. https://docs.airr-community.org/en/stable/swtools/airr_swtools_standard.html (2023).
Malone, J. et al. The Software Ontology (SWO): a resource for reproducibility in biomedical data analysis, curation and digital preservation. J. Biomed. Semant. 5, 25 (2014).
Ison, J. et al. EDAM: an ontology of bioinformatics operations, types of data and identifiers, topics and formats. Bioinformatics 29, 1325–1332 (2013).
Waltemath, D. et al. Minimum Information About a Simulation Experiment (MIASE). PLoS computational Biol. 7, e1001122 (2011).
Smith, L. P. et al. The simulation experiment description markup language (SED-ML): language specification for level 1 version 4. J. Integr. Bioinform 18, 20210021 (2021).
Bergmann, F. T. et al. COMBINE archive and OMEX format: one file to share all information to reproduce a modeling project. BMC Bioinforma. 15, 369 (2014).
Tatka L. T., Smith L. P., Hellerstein J. L., Sauro H. M. Adapting modeling and simulation credibility standards to computational systems biology. arXiv, https://doi.org/10.48550/arXiv.42301.06007 (2022).
Resource Description Framework (RDF). https://www.w3.org/RDF (2024).
Spreckelsen F., et al. Guidelines for a standardized filesystem layout for scientific data. Data 5, 43 (2020).
Greiff, V., Miho, E., Menzel, U. & Reddy, S. T. Bioinformatic and statistical analysis of adaptive immune repertoires. Trends Immunol. 36, 738–749 (2015).
Vaz, F. M., Pras-Raves, M., Bootsma, A. H. & van Kampen, A. H. Principles and practice of lipidomics. J. Inherit. Metab. Dis. 38, 41–52 (2015).
van Kampen A. H. C., Mahamune U., van Schaik B. D. C. Tools to automatic ENCORE tasks. (V1.0). AUTOMATE, https://doi.org/10.5281/zenodo.12955697 (2024).
Everything you need to learn Markdown. https://www.markdownguide.org/about (2023).
Acknowledgements
This work is supported by COSMIC which has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 765158 (AvK, RGV), by ARCAID, which has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 847551 (AvK, UM, DB), by CLOSIT, which is a collaboration project financed by the Ministry of Economic Affairs by means of the PPP Allowance made available by the Top Sector Life Sciences and Health (PM, DL), and by 3TR which has received funding from the Innovative Medicines Initiative 2 Joint Undertaking (JU) under grant agreement No 831434 (3TR). The JU receives support from the European Union’s Horizon 2020 research and innovation program and EFPIA (AvK, AJ).
Author information
Authors and Affiliations
Contributions
Av.K. and P.M. led the development of ENCORE. U.M. made major contributions to improve and test ENCORE. Av.K., U.M., and A.J. developed the Navigator. Av.K., U.M., A.J., Bv.S., D.B., D.L., M.P.R., E.W., A.D., R.G.V., and P.D. contributed to the development, evaluation, use of ENCORE, and writing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks David Gavaghan, Hunter Moseley, and Brian Schilder for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
van Kampen, A.H.C., Mahamune, U., Jongejan, A. et al. ENCORE: a practical implementation to improve reproducibility and transparency of computational research. Nat Commun 15, 8117 (2024). https://doi.org/10.1038/s41467-024-52446-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52446-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.