A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity
- PMID: 9971832
- PMCID: PMC104494
- DOI: 10.1128/JVI.73.3.2469-2480.1999
A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity
Abstract
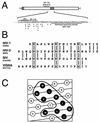
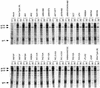
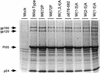
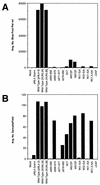
Mutations were introduced into the ectodomain of the human immunodeficiency virus type 1 (HIV-1) transmembrane envelope glycoprotein, gp41, within a region immediately adjacent to the membrane-spanning domain. This region, which is predicted to form an alpha-helix, contains highly conserved hydrophobic residues and is unusually rich in tryptophan residues. In addition, this domain overlaps the epitope of a neutralizing monoclonal antibody, 2F5, as well as the sequence corresponding to a peptide, DP-178, shown to potently neutralize virus. Site-directed mutagenesis was used to create deletions, substitutions, and insertions centered around a stretch of 17 hydrophobic and uncharged amino acids (residues 666 to 682 of the HXB2 strain of HIV-1) in order to determine the role of this region in the maturation and function of the envelope glycoprotein. Deletion of the entire stretch of 17 amino acids abrogated the ability of the envelope glycoprotein to mediate both cell-cell fusion and virus entry without affecting the normal maturation, transport, or CD4-binding ability of the protein. This phenotype was also demonstrated by substituting alanine residues for three of the five tryptophan residues within this sequence. Smaller deletions, as well as multiple amino acid substitutions, were also found to inhibit but not block cell-cell fusion. These results demonstrate the crucial role of a tryptophan-rich motif in gp41 during a post-CD4-binding step of glycoprotein-mediated fusion. The basis for the invariant nature of the tryptophans, however, appears to be at the level of glycoprotein incorporation into virions. Even the substitution of phenylalanine for a single tryptophan residue was sufficient to reduce Env incorporation and drop the efficiency of virus entry approximately 10-fold, despite the fact that the same mutation had no significant effect on syncytium formation.
Figures




Similar articles
-
Mutations of conserved glycine residues within the membrane-spanning domain of human immunodeficiency virus type 1 gp41 can inhibit membrane fusion and incorporation of Env onto virions.Jpn J Infect Dis. 2006 Apr;59(2):77-84. Jpn J Infect Dis. 2006. PMID: 16632906
-
Interhelical interactions in the gp41 core: implications for activation of HIV-1 membrane fusion.Biochemistry. 2002 Jun 11;41(23):7283-92. doi: 10.1021/bi025648y. Biochemistry. 2002. PMID: 12044159
-
Antigenic properties of the human immunodeficiency virus transmembrane glycoprotein during cell-cell fusion.J Virol. 2002 Dec;76(23):12123-34. doi: 10.1128/jvi.76.23.12123-12134.2002. J Virol. 2002. PMID: 12414953 Free PMC article.
-
Determinants of human immunodeficiency virus type 1 envelope glycoprotein oligomeric structure.J Virol. 1995 Feb;69(2):1209-18. doi: 10.1128/JVI.69.2.1209-1218.1995. J Virol. 1995. PMID: 7815497 Free PMC article.
-
The Polar Region of the HIV-1 Envelope Protein Determines Viral Fusion and Infectivity by Stabilizing the gp120-gp41 Association.J Virol. 2019 Mar 21;93(7):e02128-18. doi: 10.1128/JVI.02128-18. Print 2019 Apr 1. J Virol. 2019. PMID: 30651369 Free PMC article.
Cited by
-
A Blueprint for HIV Vaccine Discovery.Cell Host Microbe. 2012 Oct 18;12(4):396-407. doi: 10.1016/j.chom.2012.09.008. Cell Host Microbe. 2012. PMID: 23084910 Free PMC article. Review.
-
Single Amino Acid Substitution N659D in HIV-2 Envelope Glycoprotein (Env) Impairs Viral Release and Hampers BST-2 Antagonism.Viruses. 2016 Oct 14;8(10):285. doi: 10.3390/v8100285. Viruses. 2016. PMID: 27754450 Free PMC article.
-
Conformational properties of peptides corresponding to the ebolavirus GP2 membrane-proximal external region in the presence of micelle-forming surfactants and lipids.Biochemistry. 2013 May 21;52(20):3393-404. doi: 10.1021/bi400040v. Epub 2013 May 7. Biochemistry. 2013. PMID: 23650881 Free PMC article.
-
Molecular recognition of a membrane-anchored HIV-1 pan-neutralizing epitope.Commun Biol. 2022 Nov 18;5(1):1265. doi: 10.1038/s42003-022-04219-6. Commun Biol. 2022. PMID: 36400835 Free PMC article.
-
Topological analysis of the gp41 MPER on lipid bilayers relevant to the metastable HIV-1 envelope prefusion state.Proc Natl Acad Sci U S A. 2019 Nov 5;116(45):22556-22566. doi: 10.1073/pnas.1912427116. Epub 2019 Oct 17. Proc Natl Acad Sci U S A. 2019. PMID: 31624123 Free PMC article.
References
-
- Aloia R C, Curtain C C, Jensen F C. Membrane cholesterol and human immunodeficiency virus infectivity. In: Aloia R C, Curtain C C, editors. Membrane interactions of HIV: implications for pathogenesis and therapy in AIDS. Vol. 6. New York, N.Y: Wiley-Liss, Inc.; 1992. pp. 283–303.
-
- Blondelle S E, Simpkins L R, Perez-Paya E, Houghton R A. Influence of tryptophan residues on melittin’s hemolytic activity. Biochim Biophys Acta. 1993;1202:331–336. - PubMed
-
- Bosch M L, Earl P L, Fargnoli K, Picciafuoco S, Giombini F, Wong-Staal F, Franchini G. Identification of the fusion peptide of primate immunodeficiency viruses. Science. 1989;244:694–697. - PubMed
-
- Brown D, London E. Structure of detergent-resistant membrane domains: does phase separation occur in biological membranes? Biochem Biophys Res Commun. 1997;240:1–7. - PubMed
-
- Bullough P A, Hughson F M, Skehel J J, Wiley D C. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature (London) 1994;371:37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

