ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton
- PMID: 9927707
- PMCID: PMC15364
- DOI: 10.1073/pnas.96.3.1140
ARG1 (altered response to gravity) encodes a DnaJ-like protein that potentially interacts with the cytoskeleton
Abstract
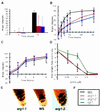
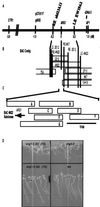
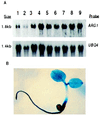
Gravitropism allows plant organs to direct their growth at a specific angle from the gravity vector, promoting upward growth for shoots and downward growth for roots. Little is known about the mechanisms underlying gravitropic signal transduction. We found that mutations in the ARG1 locus of Arabidopsis thaliana alter root and hypocotyl gravitropism without affecting phototropism, root growth responses to phytohormones or inhibitors of auxin transport, or starch accumulation. The positional cloning of ARG1 revealed a DnaJ-like protein containing a coiled-coil region homologous to coiled coils found in cytoskeleton-interacting proteins. These data suggest that ARG1 participates in a gravity-signaling process involving the cytoskeleton. A combination of Northern blot studies and analysis of ARG1-GUS fusion-reporter expression in transgenic plants demonstrated that ARG1 is expressed in all organs. Ubiquitous ARG1 expression in Arabidopsis and the identification of an ortholog in Caenorhabditis elegans suggest that ARG1 is involved in other essential processes.
Figures


Similar articles
-
The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway.Plant Physiol. 2003 Sep;133(1):100-12. doi: 10.1104/pp.103.023358. Plant Physiol. 2003. PMID: 12970478 Free PMC article.
-
A role for the TOC complex in Arabidopsis root gravitropism.Plant Physiol. 2009 Apr;149(4):1896-905. doi: 10.1104/pp.109.135301. Epub 2009 Feb 11. Plant Physiol. 2009. PMID: 19211693 Free PMC article.
-
Root gravitropism: a complex response to a simple stimulus?Trends Plant Sci. 1999 Oct;4(10):407-12. doi: 10.1016/s1360-1385(99)01472-7. Trends Plant Sci. 1999. PMID: 10498965 Review.
-
Molecular genetics of root gravitropism and waving in Arabidopsis thaliana.Gravit Space Biol Bull. 1998 May;11(2):71-8. Gravit Space Biol Bull. 1998. PMID: 11540641
-
Auxins and tropisms.J Plant Growth Regul. 2001 Sep;20(3):226-43. doi: 10.1007/s003440010027. J Plant Growth Regul. 2001. PMID: 12033223 Review.
Cited by
-
WVD2 and WDL1 modulate helical organ growth and anisotropic cell expansion in Arabidopsis.Plant Physiol. 2003 Feb;131(2):493-506. doi: 10.1104/pp.015966. Plant Physiol. 2003. PMID: 12586874 Free PMC article.
-
Isolation of New Gravitropic Mutants under Hypergravity Conditions.Front Plant Sci. 2016 Sep 29;7:1443. doi: 10.3389/fpls.2016.01443. eCollection 2016. Front Plant Sci. 2016. PMID: 27746791 Free PMC article.
-
Arabidopsis thaliana: A Model for the Study of Root and Shoot Gravitropism.Arabidopsis Book. 2002;1:e0043. doi: 10.1199/tab.0043. Epub 2002 Mar 27. Arabidopsis Book. 2002. PMID: 22303208 Free PMC article.
-
ALTERED RESPONSE TO GRAVITY is a peripheral membrane protein that modulates gravity-induced cytoplasmic alkalinization and lateral auxin transport in plant statocytes.Plant Cell. 2003 Nov;15(11):2612-25. doi: 10.1105/tpc.015560. Epub 2003 Sep 24. Plant Cell. 2003. PMID: 14507996 Free PMC article.
-
The ARG1-LIKE2 gene of Arabidopsis functions in a gravity signal transduction pathway that is genetically distinct from the PGM pathway.Plant Physiol. 2003 Sep;133(1):100-12. doi: 10.1104/pp.103.023358. Plant Physiol. 2003. PMID: 12970478 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

