Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes
- PMID: 9841678
- PMCID: PMC98952
- DOI: 10.1128/MMBR.62.4.1435-1491.1998
Protein phylogenies and signature sequences: A reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes
Abstract
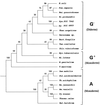
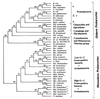
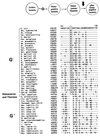
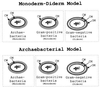
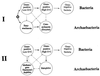
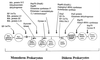
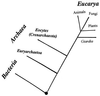
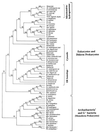
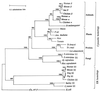
The presence of shared conserved insertion or deletions (indels) in protein sequences is a special type of signature sequence that shows considerable promise for phylogenetic inference. An alternative model of microbial evolution based on the use of indels of conserved proteins and the morphological features of prokaryotic organisms is proposed. In this model, extant archaebacteria and gram-positive bacteria, which have a simple, single-layered cell wall structure, are termed monoderm prokaryotes. They are believed to be descended from the most primitive organisms. Evidence from indels supports the view that the archaebacteria probably evolved from gram-positive bacteria, and I suggest that this evolution occurred in response to antibiotic selection pressures. Evidence is presented that diderm prokaryotes (i.e., gram-negative bacteria), which have a bilayered cell wall, are derived from monoderm prokaryotes. Signature sequences in different proteins provide a means to define a number of different taxa within prokaryotes (namely, low G+C and high G+C gram-positive, Deinococcus-Thermus, cyanobacteria, chlamydia-cytophaga related, and two different groups of Proteobacteria) and to indicate how they evolved from a common ancestor. Based on phylogenetic information from indels in different protein sequences, it is hypothesized that all eukaryotes, including amitochondriate and aplastidic organisms, received major gene contributions from both an archaebacterium and a gram-negative eubacterium. In this model, the ancestral eukaryotic cell is a chimera that resulted from a unique fusion event between the two separate groups of prokaryotes followed by integration of their genomes.
Figures























Similar articles
-
What are archaebacteria: life's third domain or monoderm prokaryotes related to gram-positive bacteria? A new proposal for the classification of prokaryotic organisms.Mol Microbiol. 1998 Aug;29(3):695-707. doi: 10.1046/j.1365-2958.1998.00978.x. Mol Microbiol. 1998. PMID: 9723910 Review.
-
The natural evolutionary relationships among prokaryotes.Crit Rev Microbiol. 2000;26(2):111-31. doi: 10.1080/10408410091154219. Crit Rev Microbiol. 2000. PMID: 10890353 Review.
-
Phylogenetic analysis of 70 kD heat shock protein sequences suggests a chimeric origin for the eukaryotic cell nucleus.Curr Biol. 1994 Dec 1;4(12):1104-14. doi: 10.1016/s0960-9822(00)00249-9. Curr Biol. 1994. PMID: 7704574
-
Protein phylogenies and signature sequences: evolutionary relationships within prokaryotes and between prokaryotes and eukaryotes.Antonie Van Leeuwenhoek. 1997 Jul;72(1):49-61. doi: 10.1023/a:1000278224701. Antonie Van Leeuwenhoek. 1997. PMID: 9296263
-
Life's third domain (Archaea): an established fact or an endangered paradigm?Theor Popul Biol. 1998 Oct;54(2):91-104. doi: 10.1006/tpbi.1998.1376. Theor Popul Biol. 1998. PMID: 9733652 Review.
Cited by
-
Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria.Microbiol Mol Biol Rev. 2012 Mar;76(1):66-112. doi: 10.1128/MMBR.05011-11. Microbiol Mol Biol Rev. 2012. PMID: 22390973 Free PMC article. Review.
-
Influence of rhizobacterial volatiles on the root system architecture and the production and allocation of biomass in the model grass Brachypodium distachyon (L.) P. Beauv.BMC Plant Biol. 2015 Aug 12;15:195. doi: 10.1186/s12870-015-0585-3. BMC Plant Biol. 2015. PMID: 26264238 Free PMC article.
-
Development of the primer set for the detection of the hsp60 gene in Trichophyton mentagrophytes cDNA.Folia Microbiol (Praha). 1999;44(4):401-5. doi: 10.1007/BF02903713. Folia Microbiol (Praha). 1999. PMID: 10983236
-
A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history.Genome Res. 2002 Jul;12(7):1080-90. doi: 10.1101/gr.187002. Genome Res. 2002. PMID: 12097345 Free PMC article.
-
Evolution of viruses by acquisition of cellular RNA or DNA nucleotide sequences and genes: an introduction.Virus Genes. 2000;21(1-2):7-12. Virus Genes. 2000. PMID: 11022785 Review.
References
-
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J D. Molecular biology of the cell. New York, N.Y: Garland Publishing, Inc.; 1994.
-
- Allsopp A. Phylogenetic relationships of the procaryota and the origin of the eucaryotic cell. New Phytol. 1969;68:591–612.
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

